-
spaCRT: saddlepoint approximation-based conditional randomization test
This note is for Niu, Z., Choudhury, J. R., & Katsevich, E. (2024). Computationally efficient and statistically accurate conditional independence testing with spaCRT (No. arXiv:2407.08911; Version 1). arXiv. https://doi.org/10.48550/arXiv.2407.08911
-
scDRS: single-cell disease relevance score
This note is for Zhang, M. J., Hou, K., Dey, K. K., Sakaue, S., Jagadeesh, K. A., Weinand, K., Taychameekiatchai, A., Rao, P., Pisco, A. O., Zou, J., Wang, B., Gandal, M., Raychaudhuri, S., Pasaniuc, B., & Price, A. L. (2022). Polygenic enrichment distinguishes disease associations of individual cells in single-cell RNA-seq data. Nature Genetics, 54(10), 1572–1580. https://doi.org/10.1038/s41588-022-01167-z
-
Conformal Prediction for Single-cell Spatial Transcriptomics
This note is for Sun, E. D., Ma, R., Navarro Negredo, P., Brunet, A., & Zou, J. (2024). TISSUE: Uncertainty-calibrated prediction of single-cell spatial transcriptomics improves downstream analyses. Nature Methods, 21(3), 444–454.
-
Niche DE
This note is for Mason, K., Sathe, A., Hess, P. R., Rong, J., Wu, C.-Y., Furth, E., Susztak, K., Levinsohn, J., Ji, H. P., & Zhang, N. (2024). Niche-DE: Niche-differential gene expression analysis in spatial transcriptomics data identifies context-dependent cell-cell interactions. Genome Biology, 25(1), 14.
-
Conditional Independence Test in Single-cell Multiomics
This note is for Boyeau, P., Bates, S., Ergen, C., Jordan, M. I., & Yosef, N. (2023). Calibrated Identification of Feature Dependencies in Single-cell Multiomics.
-
Comparisons of transformations for single-cell RNA-seq data
This post is for Ahlmann-Eltze, C., & Huber, W. (2023). Comparison of transformations for single-cell RNA-seq data. Nature Methods, 20(5), 665–672.
-
sctransform: Normalization using Regularized Negative Binomial Regression
The note is for Hafemeister, C., & Satija, R. (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biology, 20(1), 296.
-
One-way Matching with Low Rank
This post is for Chen, Shuxiao, Sizun Jiang, Zongming Ma, Garry P. Nolan, and Bokai Zhu. “One-Way Matching of Datasets with Low Rank Signals.” arXiv, October 3, 2022.
-
CountSplit for scRNA Data
The post is for Neufeld, Anna, Lucy L Gao, Joshua Popp, Alexis Battle, and Daniela Witten. “Inference after Latent Variable Estimation for Single-cell RNA Sequencing Data.” Biostatistics, December 13, 2022, kxac047.
-
Uncertainty of Pseudotime Trajectory
This post is for Tenha, Lovemore, and Mingzhou Song. “Statistical Evidence for the Presence of Trajectory in Single-cell Data.” BMC Bioinformatics 23, no. Suppl 8 (August 16, 2022): 340.
-
ClusterDE: a post-clustering DE method
This post is for Song, Dongyuan, Kexin Li, Xinzhou Ge, and Jingyi Jessica Li. “ClusterDE: A Post-Clustering Differential Expression (DE) Method Robust to False-Positive Inflation Caused by Double Dipping,” 2023
-
scHOT: Investigate higher-order interactions in single-cell data
This note is for Ghazanfar, Shila, Yingxin Lin, Xianbin Su, David Ming Lin, Ellis Patrick, Ze-Guang Han, John C. Marioni, and Jean Yee Hwa Yang. “Investigating Higher-Order Interactions in Single-cell Data with scHOT.” Nature Methods 17, no. 8 (August 2020): 799–806.
-
Lamian: Differential Pseudotime Analysis
This note is for Hou, W., Ji, Z., Chen, Z., Wherry, E. J., Hicks, S. C., & Ji, H. (2021). A statistical framework for differential pseudotime analysis with multiple single-cell RNA-seq samples (p. 2021.07.10.451910). bioRxiv.
-
condiments: Trajectory Inference across Multiple Conditions
The note is for Van den Berge, K., Roux de Bézieux, H., Street, K., Saelens, W., Cannoodt, R., Saeys, Y., Dudoit, S., & Clement, L. (2020). Trajectory-based differential expression analysis for single-cell sequencing data. Nature Communications, 11(1), Article 1.
-
tradeSeq: Trajectory-based differential expression analysis for single-cell sequencing data

This post is for Van den Berge, K., Roux de Bézieux, H., Street, K., Saelens, W., Cannoodt, R., Saeys, Y., Dudoit, S., & Clement, L. (2020). Trajectory-based differential expression analysis for single-cell sequencing data. Nature Communications, 11(1), Article 1.
-
PseudotimeDE: Differential Gene Expression along Cell Pseudotime
The note is for Song, D., & Li, J. J. (2021). PseudotimeDE: Inference of differential gene expression along cell pseudotime with well-calibrated p-values from single-cell RNA sequencing data. Genome Biology, 22(1), 124.
-
scMDC: Single-cell Multi-omics Data Clustering Analysis
This post is for Lin, X., Tian, T., Wei, Z., & Hakonarson, H. (2022). Clustering of single-cell multi-omics data with a multimodal deep learning method. Nature Communications, 13(1), Article 1.
-
Single Cell Generative Pre-trained Transformer

This post is for Cui, H., Wang, C., Maan, H., & Wang, B. (2023). scGPT: Towards Building a Foundation Model for Single-cell Multi-omics Using Generative AI (p. 2023.04.30.538439). bioRxiv.
-
Deep Generative Modeling for Single-cell Transcriptomics

The post is for Lopez, R., Regier, J., Cole, M. B., Jordan, M. I., & Yosef, N. (2018). Deep generative modeling for single-cell transcriptomics. Nature Methods, 15(12), Article 12.
-
scDesign3: A Single-cell Simulator
This note is based on Jingyi Jessica Li’s talk on Song, D., Wang, Q., Yan, G., Liu, T., & Li, J. J. (2022). A unified framework of realistic in silico data generation and statistical model inference for single-cell and spatial omics (p. 2022.09.20.508796). bioRxiv.
-
Single-cell Graph Neural Network

This note is for Prof. Dong Xu’s talk on Wang, J., Ma, A., Chang, Y., Gong, J., Jiang, Y., Qi, R., Wang, C., Fu, H., Ma, Q., & Xu, D. (2021). ScGNN is a novel graph neural network framework for single-cell RNA-Seq analyses. Nature Communications, 12(1), Article 1.
-
An objective comparison of cell-tracking algorithms
This note is for Ulman, V., Maška, M., Magnusson, K. E. G., Ronneberger, O., Haubold, C., Harder, N., Matula, P., Matula, P., Svoboda, D., Radojevic, M., Smal, I., Rohr, K., Jaldén, J., Blau, H. M., Dzyubachyk, O., Lelieveldt, B., Xiao, P., Li, Y., Cho, S.-Y., … Ortiz-de-Solorzano, C. (2017). An objective comparison of cell-tracking algorithms. Nature Methods, 14(12), 1141–1152.
-
Eleven Challengs in Single Cell Data Science
This note is for Lähnemann, D., Köster, J., Szczurek, E., McCarthy, D. J., Hicks, S. C., Robinson, M. D., Vallejos, C. A., Campbell, K. R., Beerenwinkel, N., Mahfouz, A., Pinello, L., Skums, P., Stamatakis, A., Attolini, C. S.-O., Aparicio, S., Baaijens, J., Balvert, M., Barbanson, B. de, Cappuccio, A., … Schönhuth, A. (2020). Eleven grand challenges in single-cell data science. Genome Biology, 21(1), 31.
-
DNA copy number profiling: from bulk tissue to single cells
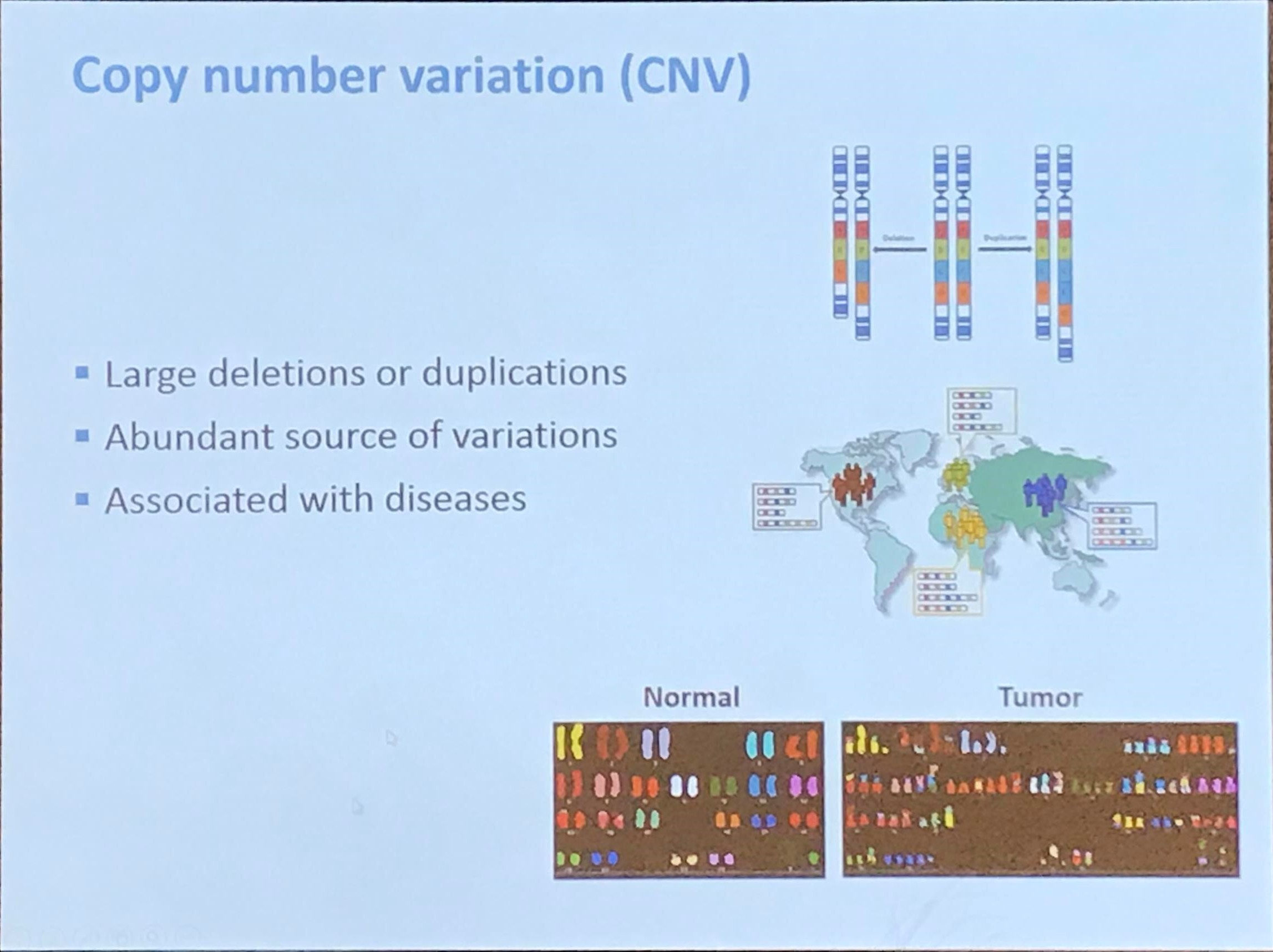
This post is based on the talk given by Yuchao Jiang at the 11th ICSA International Conference on Dec. 20th, 2019.
-
Model-based Approach for Joint Analysis of Single-cell data

This post is based on Lin, Z., Zamanighomi, M., Daley, T., Ma, S., & Wong, W. H. (2020). Model-Based Approach to the Joint Analysis of Single-cell Data on Chromatin Accessibility and Gene Expression. Statistical Science, 35(1), 2–13.





