Lamian: Differential Pseudotime Analysis
Posted on (Update: )
the majority of the methods were designed to infer gene expression changes along the reconstructed trajectory within one biological sample.
However, scRNA-seq experiments today standardly generate data with multiple biological samples across multiple conditions
Changes in pseudotemporal trajectories across conditions can occur in multiple ways, including
- topological differences
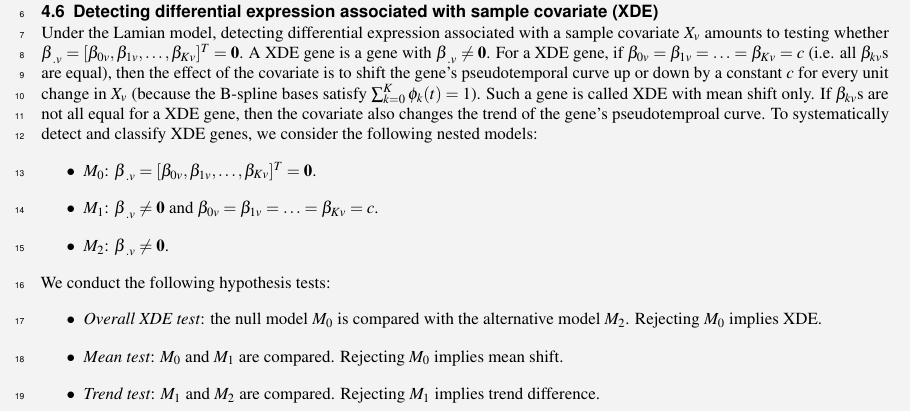
- changes in the proportion (or density or abundance) of cells along a cell lineage across conditions
- changes in the gene expression itself along pseudotime across conditions
Given scRNA-seq data from multiple biological samples with known covariates, such as age, sex, sample type, disease status, Lamian can be used to
- construct pseudotemporal trajectories and evaluate the uncertainty of the topologies
- evaluate differential changes in the topological structure associated with sample covariates
- describe how gene expression and cell density change along the pseudotime
- characterize how sample covariates modifies the pseudotemporal dynamics of gene expression and cell density
Lamian accounts for variability across biological samples. And hence Lamian is able to more appropriately control the false discovery rate (FDR) when analyzing multi-sample data.

Trajectory Construction and Uncertainty Evaluation
construct the trajectory using a cluster-based minimum spanning tree (cMST) approach
- K-means clustering is applied to cluster cells based on the top principal components (PCs) of log2-transformed library-size-normalized gene expression profile
- trajectories are then inferred as in TSCAN by a MST that treats cluster center as nodes
- number of PCs and cell cluster number are both determined using an elbow method in TSCAN
- the origin of the pseudotime is specified by users based on marker gene expression
for each of the branch, characterize its uncertainty using its detection rate in 10000 bootstrapn samples
- whether each branch in the original data is also identified in the bootstrap sample by performing pairwise comparison of branches between the original and bootstrap data
- for a pair of branches, use the Jaccard index to evaluate their overlap
- to determine the cutoff of Jaccard index, a null distribution of Jaccard index is constructed by evaluating the overlap between the cells in the branch and a randomly sampled set of cells with the cell number matching those in the branch for 1000 times