-
Generalized Higher Criticism in GWAS
This note is for Barnett, I., Mukherjee, R., & Lin, X. (2017). The Generalized Higher Criticism for Testing SNP-Set Effects in Genetic Association Studies. Journal of the American Statistical Association, 112(517), 64–76.
-
GhostKnockoffs: Only Summary Statistics
This note is for Chen, Z., He, Z., Chu, B. B., Gu, J., Morrison, T., Sabatti, C., & Candès, E. (2024). Controlled Variable Selection from Summary Statistics Only? A Solution via GhostKnockoffs and Penalized Regression (arXiv:2402.12724). arXiv.
-
Time-varying Group Sparse Additive Model for GWAS
This post is for Marchetti-Bowick, M., Yin, J., Howrylak, J. A., & Xing, E. P. (2016). A time-varying group sparse additive model for genome-wide association studies of dynamic complex traits. Bioinformatics, 32(19), 2903–2910.
-
fGWAS: Dynamic Model for GWAS
The note is for Das, K., Li, J., Wang, Z., Tong, C., Fu, G., Li, Y., Xu, M., Ahn, K., Mauger, D., Li, R., & Wu, R. (2011). A dynamic model for genome-wide association studies. Human Genetics, 129(6), 629–639.
-
GWAS of Longitudinal Trajectories at Biobank Scale

This post is for Ko, S., German, C. A., Jensen, A., Shen, J., Wang, A., Mehrotra, D. V., Sun, Y. V., Sinsheimer, J. S., Zhou, H., & Zhou, J. J. (2022). GWAS of longitudinal trajectories at biobank scale. The American Journal of Human Genetics, 109(3), 433–445.
-
Tutorial on Polygenic Risk Score
This note is based on Choi, S. W., Mak, T. S.-H., & O’Reilly, P. F. (2020). Tutorial: A guide to performing polygenic risk score analyses. Nature Protocols, 15(9), Article 9.
-
LD Score Regression
This note is for Bulik-Sullivan, B. K., Loh, P.-R., Finucane, H. K., Ripke, S., Yang, J., Patterson, N., Daly, M. J., Price, A. L., & Neale, B. M. (2015). LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature Genetics, 47(3), 291–295.
-
Joint Local False Discovery Rate in GWAS
This note is for Jiang, W., & Yu, W. (2017). Controlling the joint local false discovery rate is more powerful than meta-analysis methods in joint analysis of summary statistics from multiple genome-wide association studies. Bioinformatics, 33(4), 500–507.
-
Debiased Inverse-Variance Weighted Estimator in Mendelian Randomization
This post is for the talk at Yale given by Prof. Ting Ye based on the paper Ye, T., Shao, J., & Kang, H. (2020). Debiased Inverse-Variance Weighted Estimator in Two-Sample Summary-Data Mendelian Randomization (arXiv:1911.09802). arXiv.
-
Quantitative Genetics

This post is based on the Pao-Lu Hsu Award Lecture given by Prof. Hongyu Zhao at the 11th ICSA International Conference on Dec. 21th, 2019.
-
Controlling bias and inflation in EWAS/TWAS
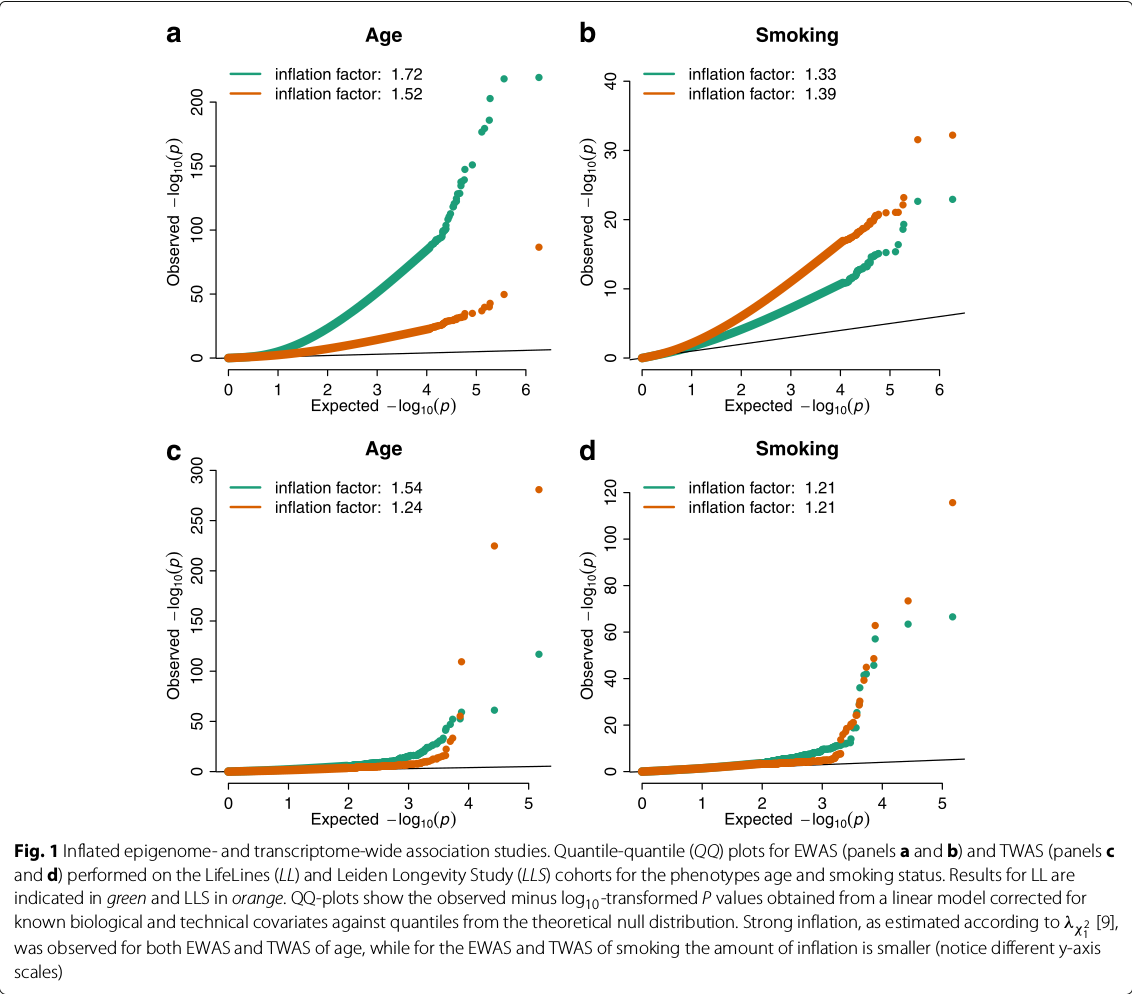
The post is based on the BIOS Consortium, van Iterson, M., van Zwet, E. W., & Heijmans, B. T. (2017). Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genome Biology, 18(1), 19.
-
Genetic Relatedness in High-Dimensional Linear Models
This post is based on Guo, Z., Wang, W., Cai, T. T., & Li, H. (2019). Optimal Estimation of Genetic Relatedness in High-Dimensional Linear Models. Journal of the American Statistical Association, 114(525), 358–369.


