Machine Learning for Multi-omics Data
Posted on (Update: )
This note is based on Cai, Z., Poulos, R. C., Liu, J., & Zhong, Q. (2022). Machine learning for multi-omics data integration in cancer. IScience, 25(2), 103798.
Multi-omics data analysis is an important aspect of cancer molecular biology studies and has led to ground-breaking discoveries.
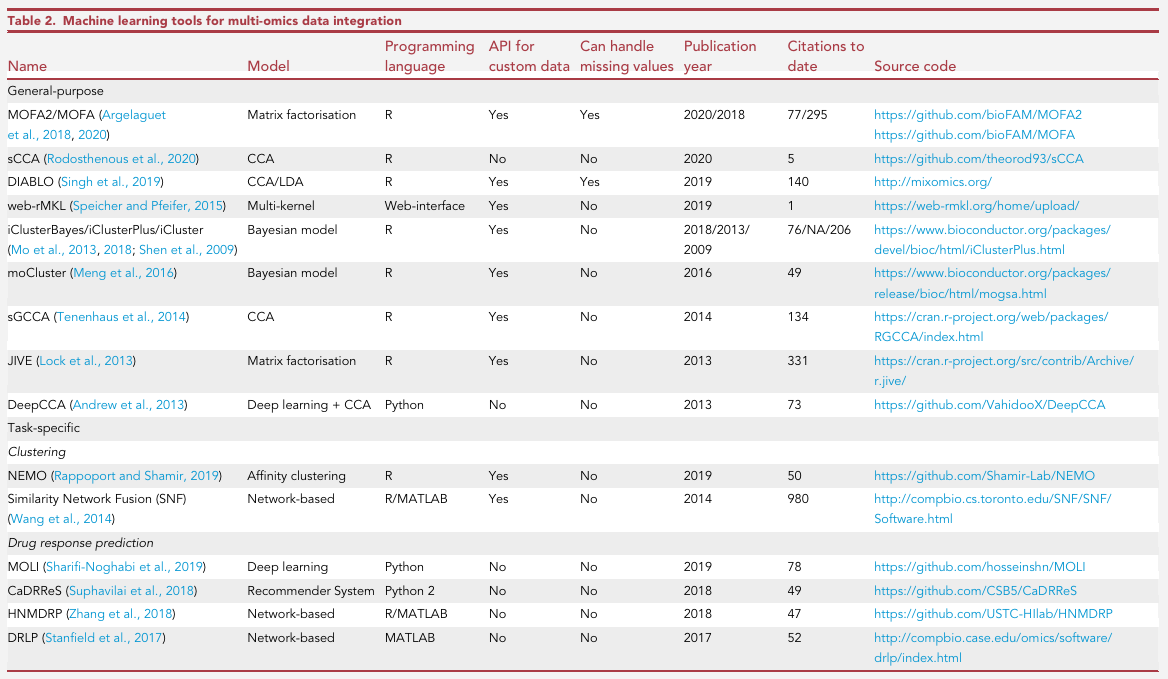
The paper review machine learning tools categorized as either general-purpose or task-specific, covering both supervised and unsupervised learning for integrative analysis of multi-omics data.
Benchmark the performance of five machine learning approaches using data from the Cancer Cell Line Encyclopedia, reporting
- cancer type classification
- mean absolute error on drug response prediction
- evaluate runtime efficiency
promote the development of novel machine learning methodologies for data integration, which will be essential for drug discovery, clinical trial design, and personalized treatments
The discipline in molecular biology that aims for the collective characterization and quantification of the genome, transcriptome, and proteome, to influence the structure, function, and dynamics of a biological sample is termed omics.
Single-omics datasets have failed to produce the revolution in cancer treatment.
Developing a holistic view of cancer behavior and identification of new therapeutic vulnerabilities may only be possible through multi-omics analysis, which has become an area of increasing interest in cancer research over the last decades.
strategies are required to systematically integrate heterogeneous multi-omics datasets to deliver actionable results that may advance biological sciences and eventually translate into clinical practice.
published multi-omics data
- TCGA: initiated by the National Cancer Institute in 2006, which generated multi-omics data for more than 20,000 tumors spanning 33 cancer types
- ICGC (International Cancer Genome Consortium): initiated by multiple countries as a collaborative program, which incorporates some projects from TCGA and features a user-friendly online analysis interface
- COSMIC (The Catalog of Somatic Mutations In Cancer): led by the Wellcome Sanger Institute and curates multi-omics data for both cancer cell lines and tumors
- DepMap (The Cancer Dependency Map): a platform similar to COSMIC developed by the Broad Institute, which provides genome-wide CRISPR-Cas9 knockout screens with comprehensive multi-omics molecular characterization of cell lines and the corresponding drug screens.

three common strategies for multi-omics data integration:

contribution of this review:

methods:
benchmarking:
