NGS for NGS
Posted on (Update: )
This post is based on the talk, Next-Generation Statistical Methods for Association Analysis of Now-Generation Sequencing Studies, given by Dr. Xiang Zhan at the Department of Statistics and Data Science, Southern University of Science and Technology on Jan. 05, 2020.
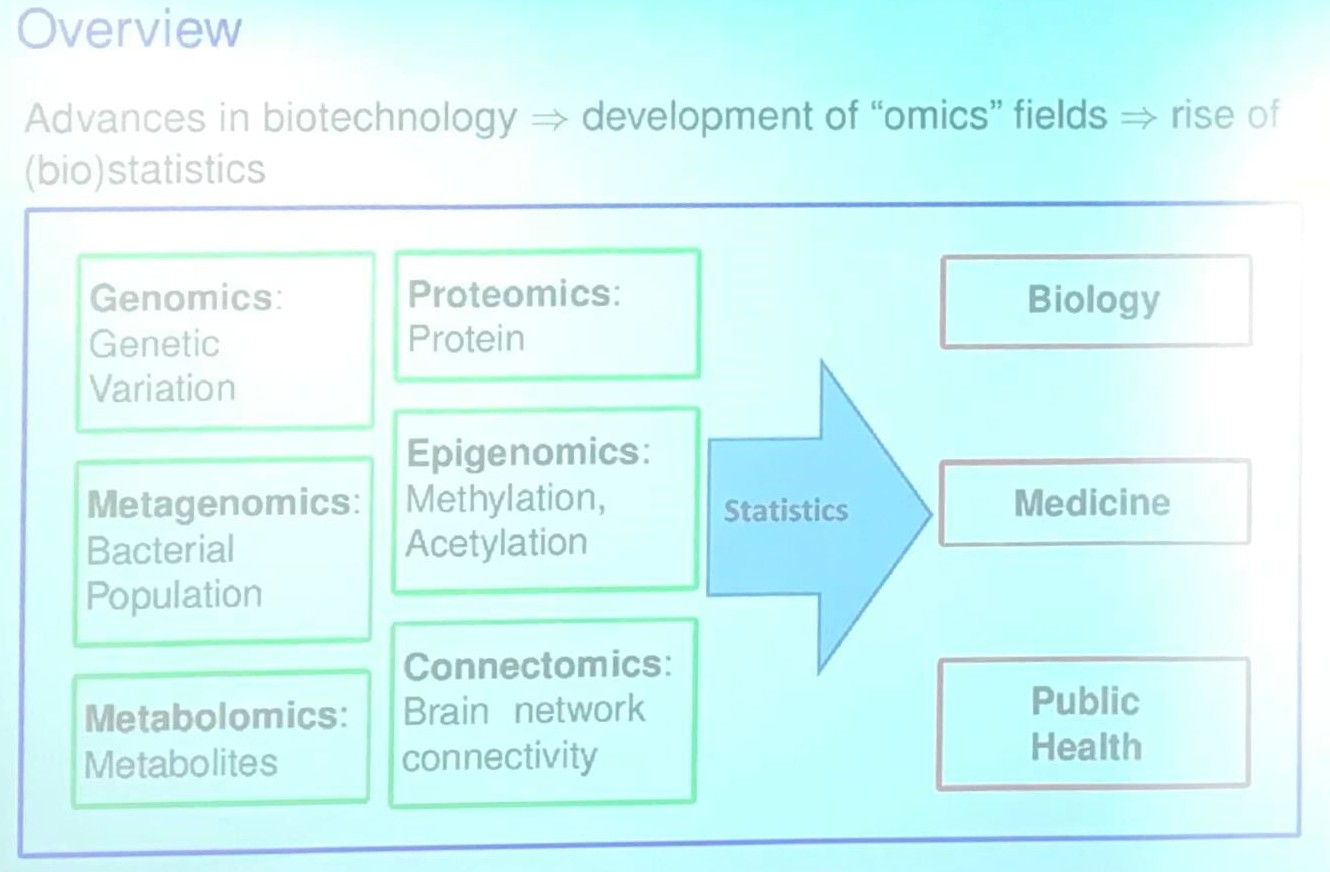
Project I: Statistical Methods for Set-based Omics Association Analysis
Goal: Test
\[H_0: \beta_1 = \cdots = \beta_p = 0\]in
\[y=X\alpha + Z_1\beta_1 + \cdots + Z_p\beta_p + \epsilon\,.\]KAT (Kernel-based Association Test)
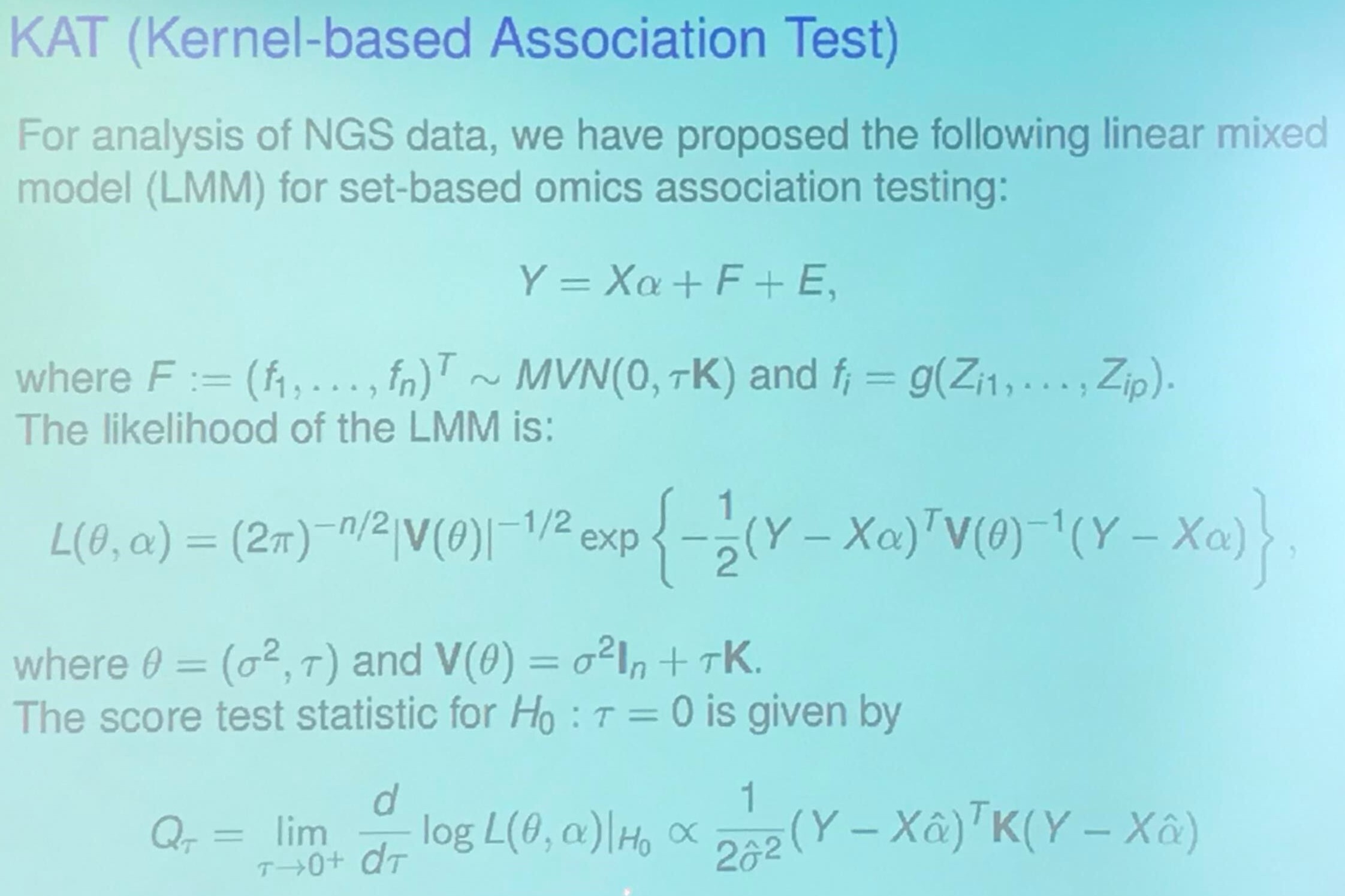
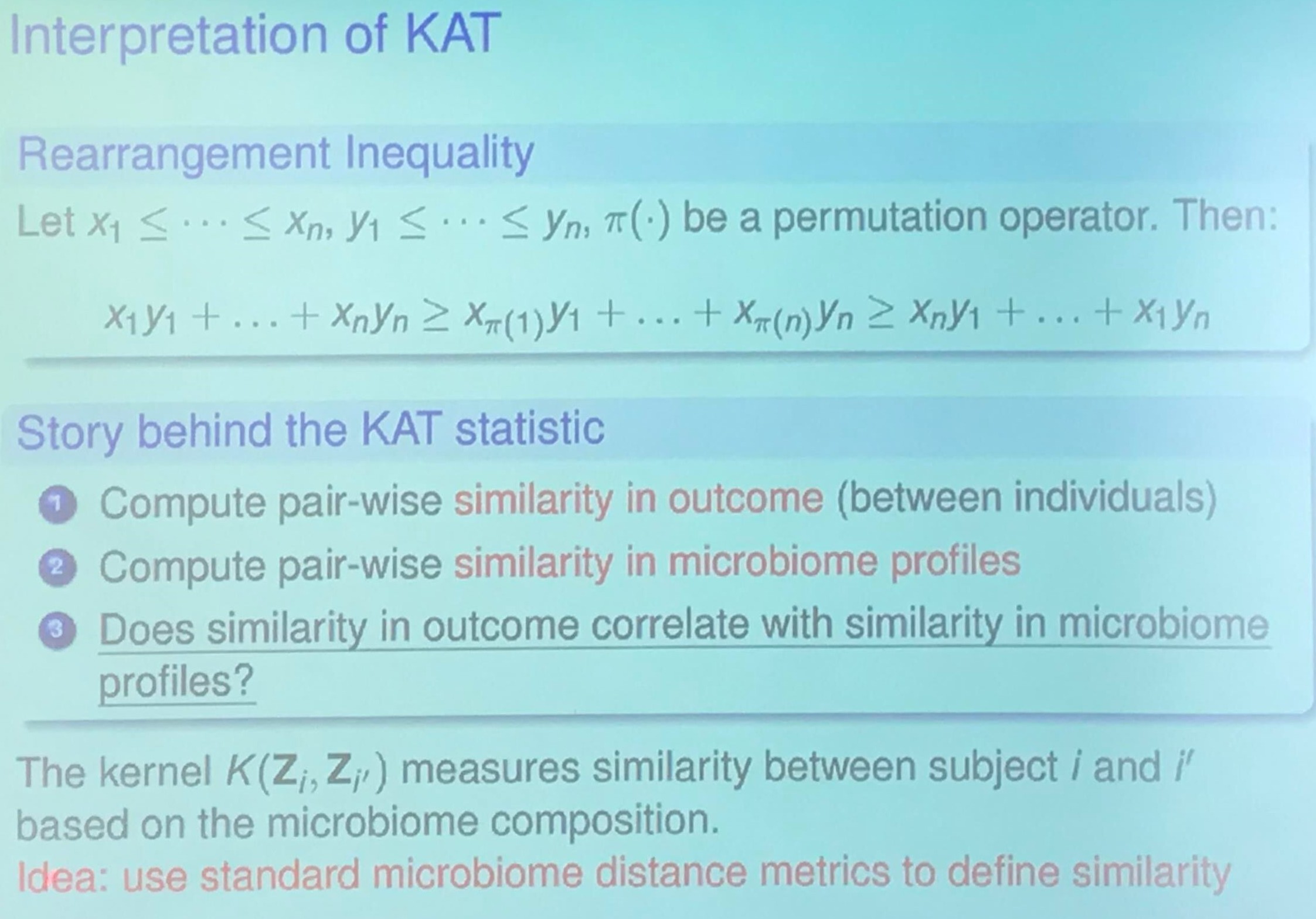
I am confused about the relationship between Rearrangement Inequality and KAT statistic. My guess is that it wants to show higher score implies higher correlation.
Microbiome Data
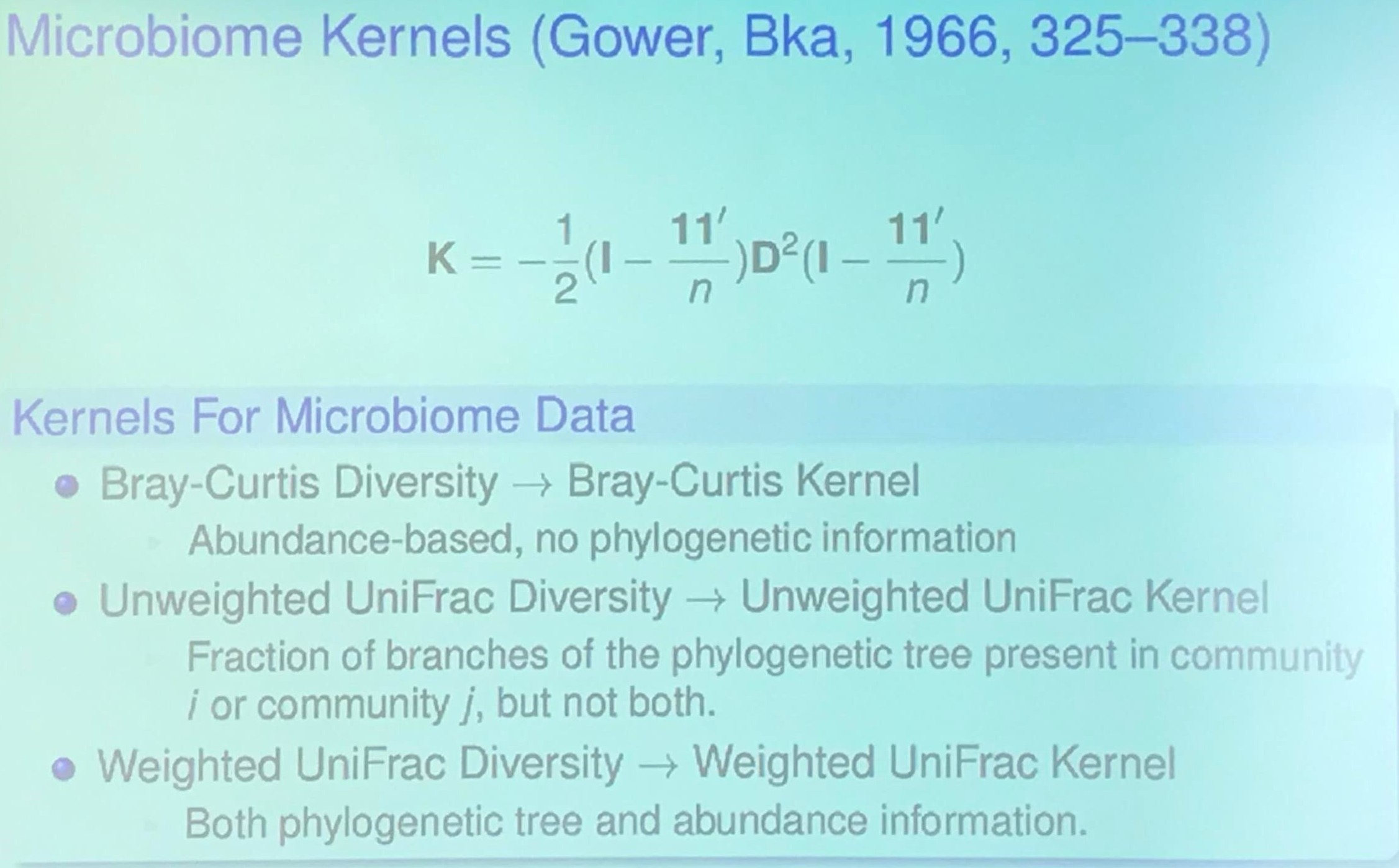
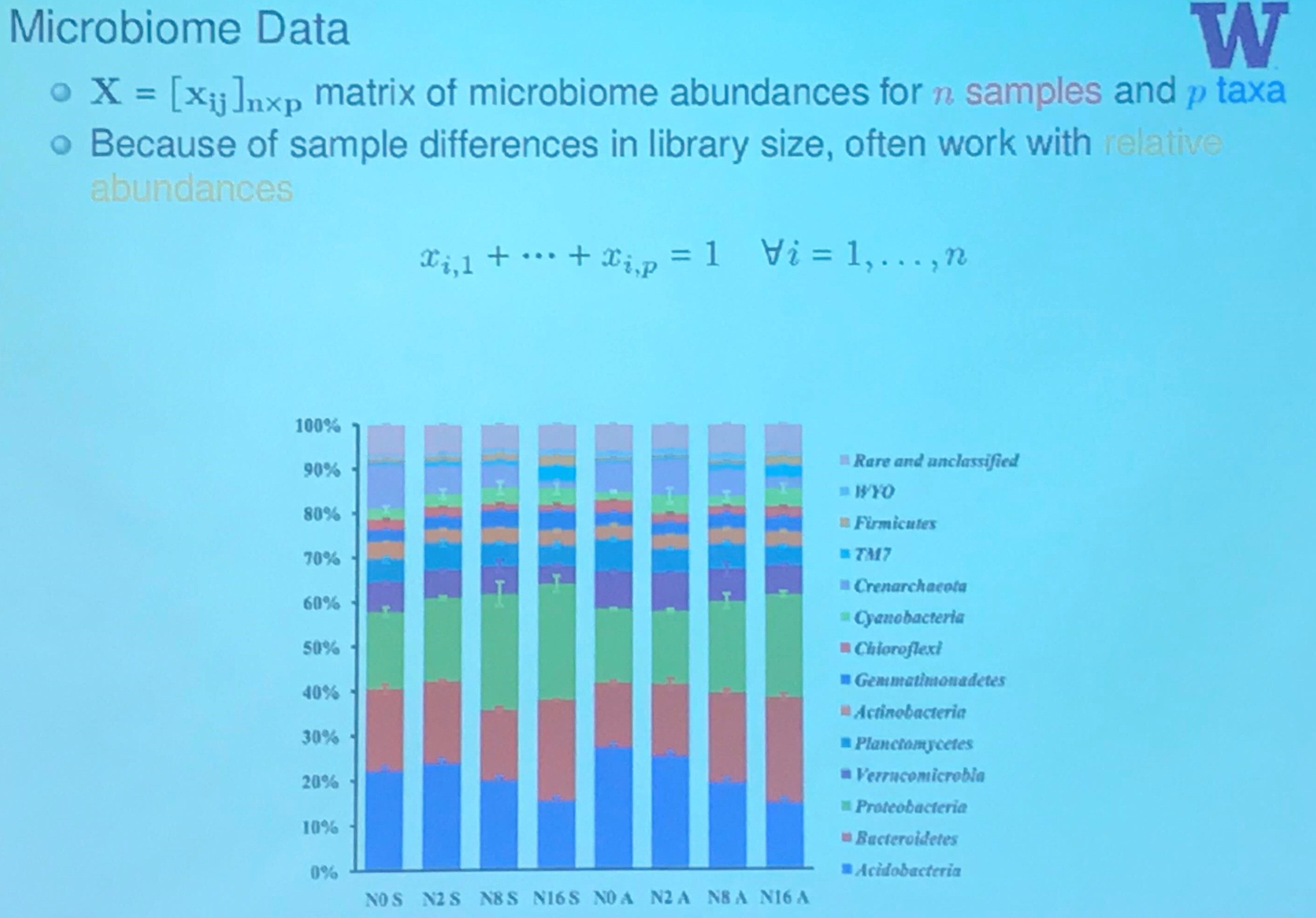 (Actually, this slide is from another speak in that conference, I will have a post later.)
(Actually, this slide is from another speak in that conference, I will have a post later.)

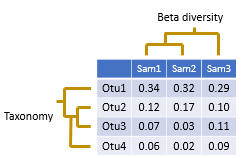
An OTU table is a matrix that gives the number of reads per sample per OTU. One entry in the table is usually a number of reads, also called a “count”, or a frequency in the range 0.0 to 1.0.
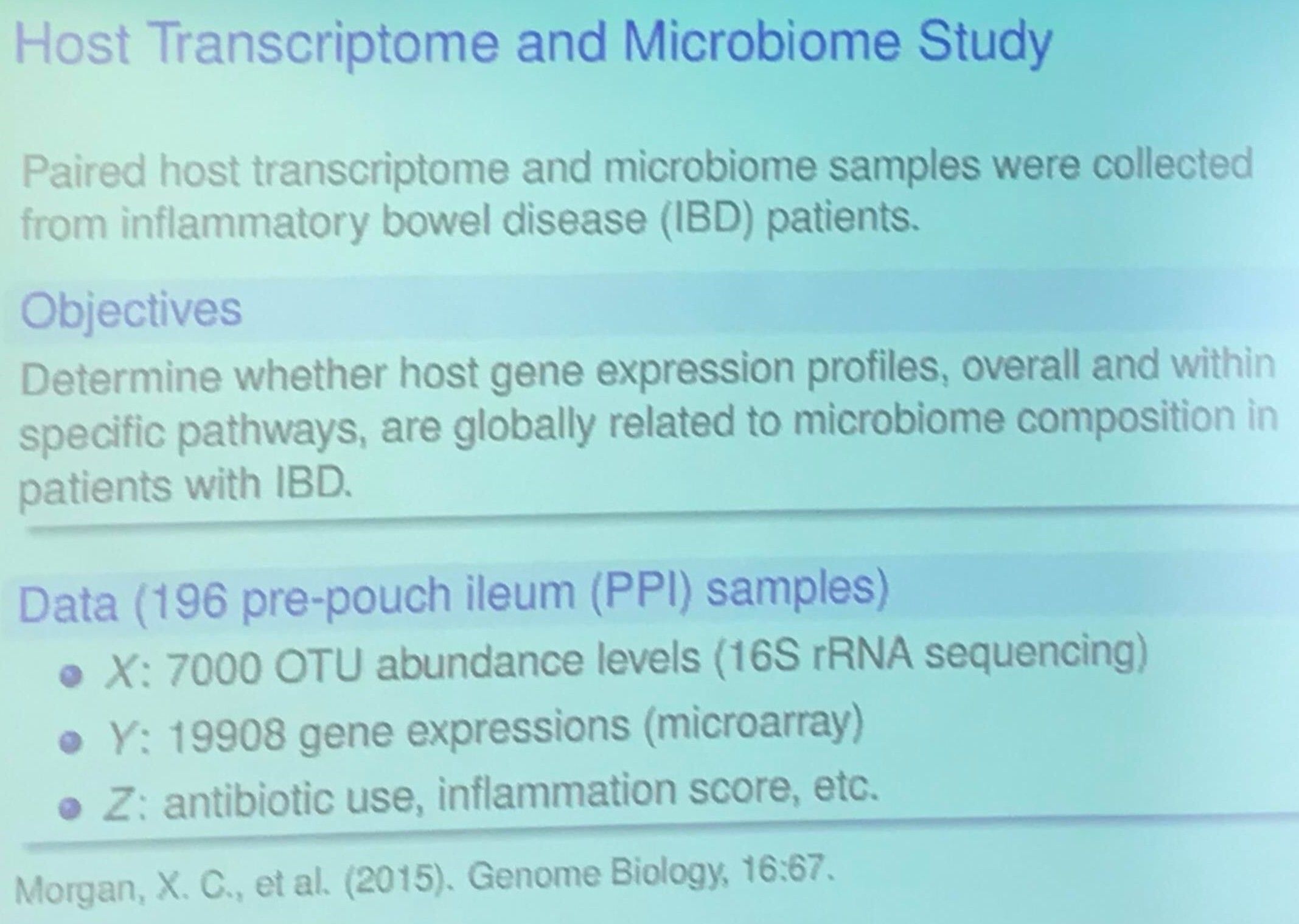
Host Transcriptome and Microbiome Study (TODO)
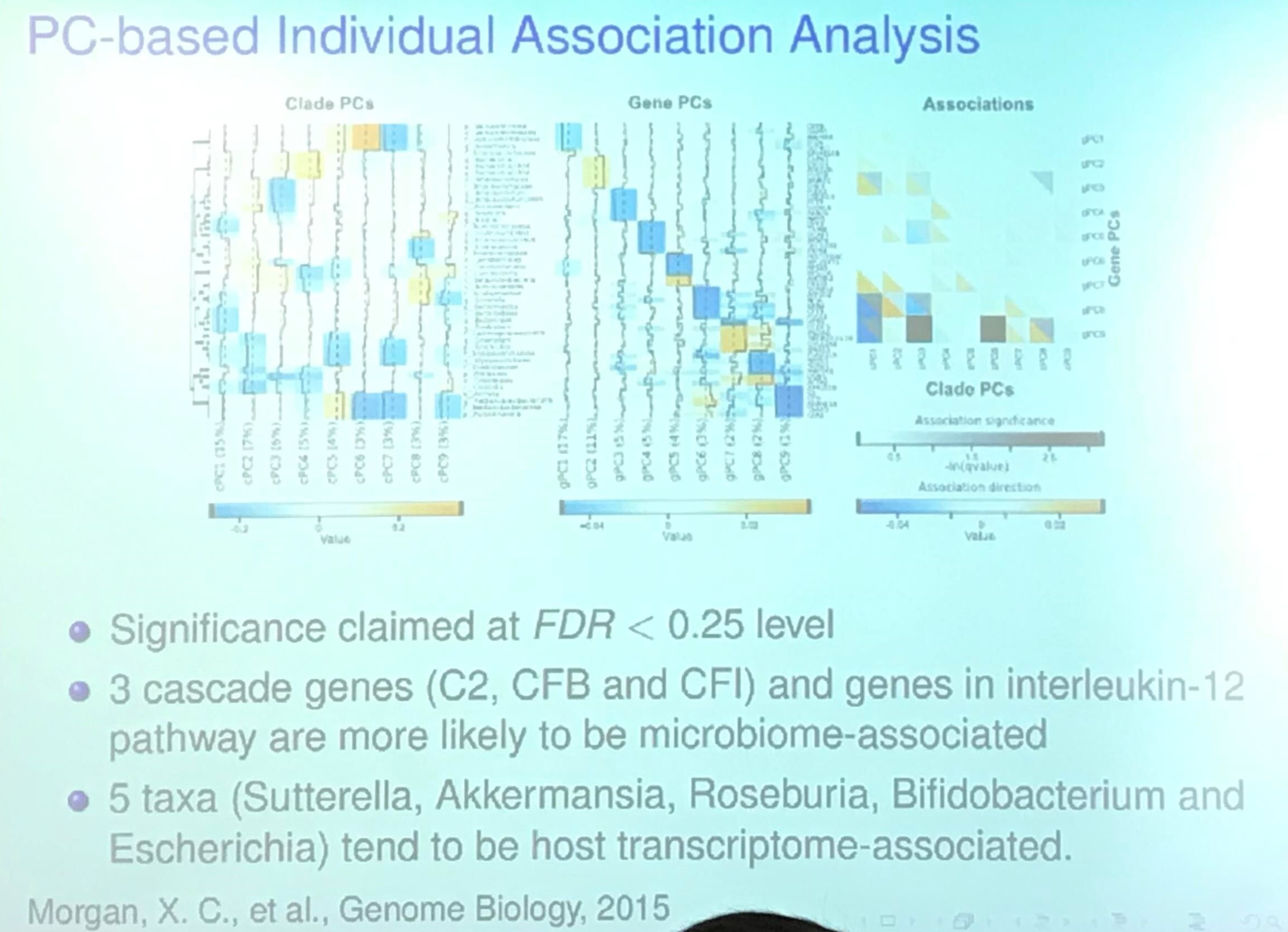
Project II: Variable Selection Methods for Prioritizing Omics Association Signals
Goal: Identify $\{j:\beta_j\neq 0, j=1,\ldots, p\}$ in
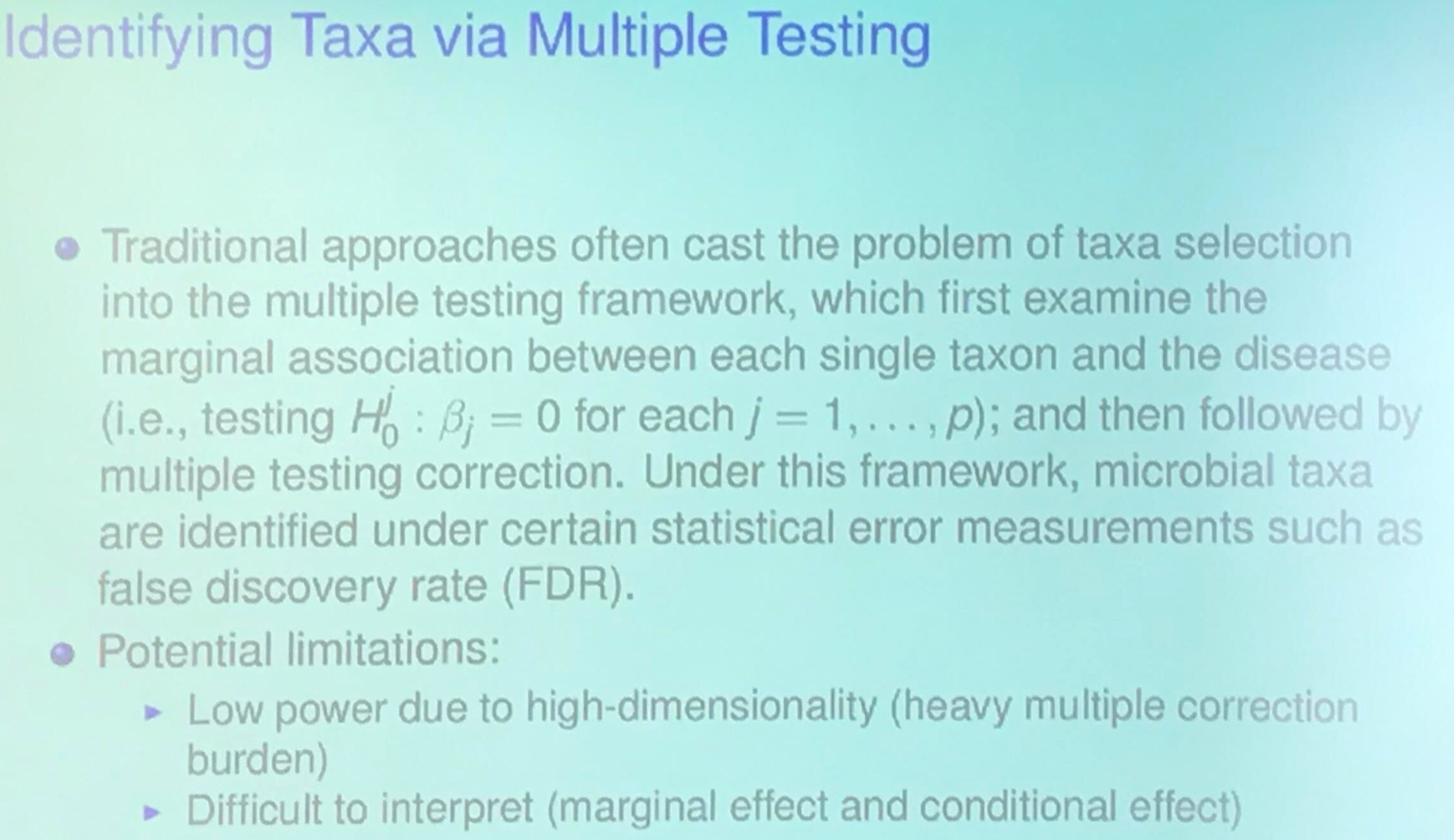
\[y = \beta_1X_1 + \cdots +\beta_pX_p + \epsilon\,.\]Identifying Taxa via Multiple Testing
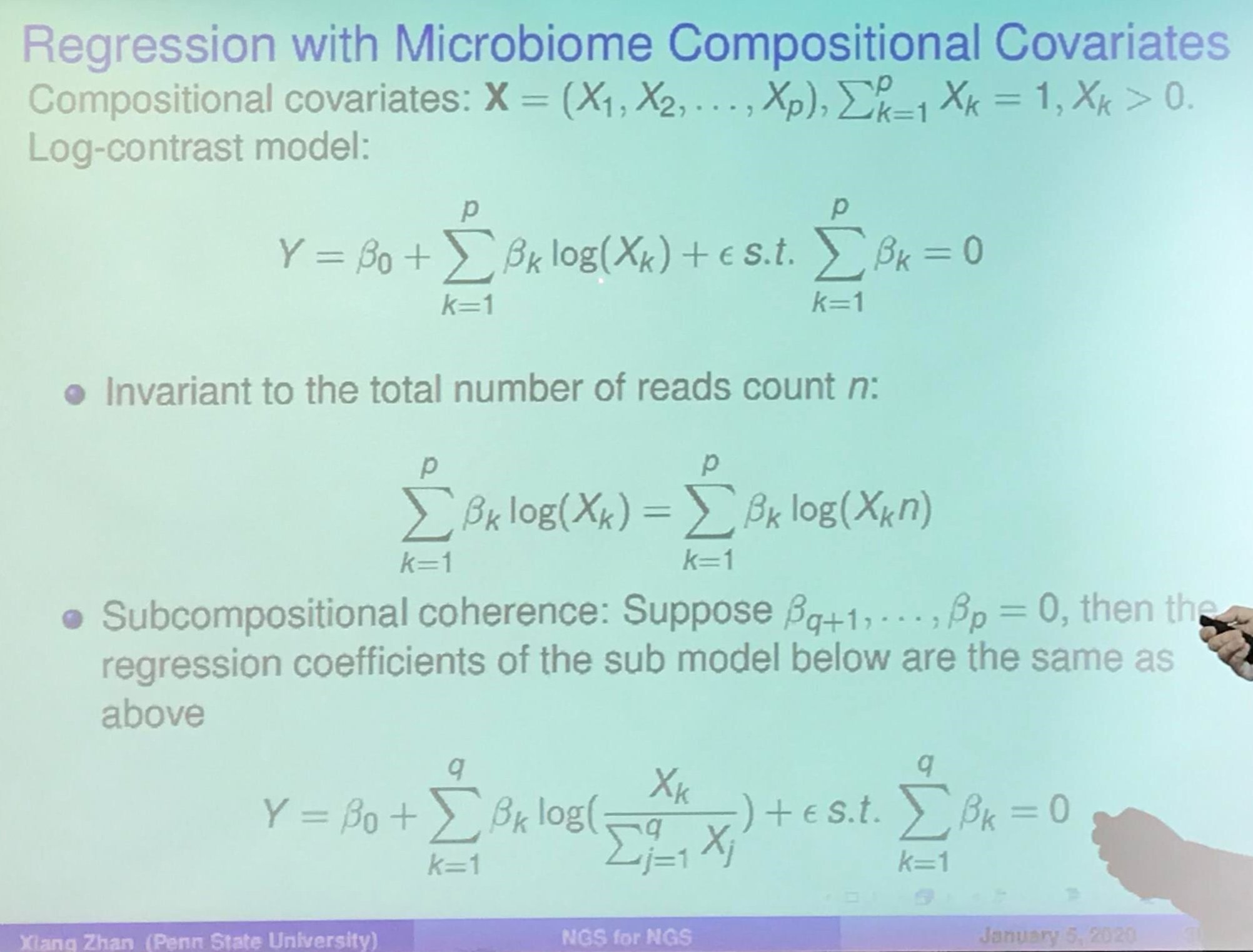
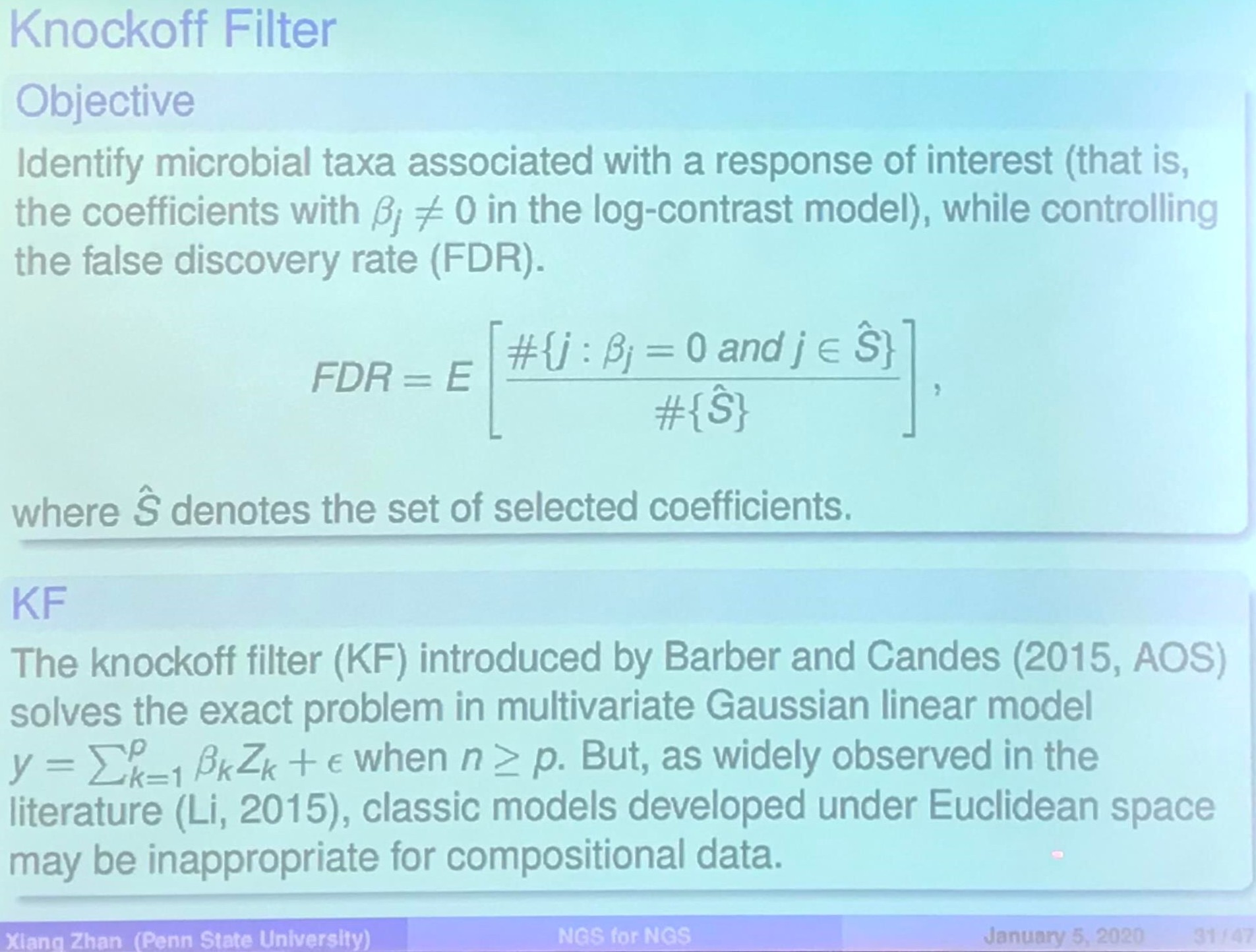
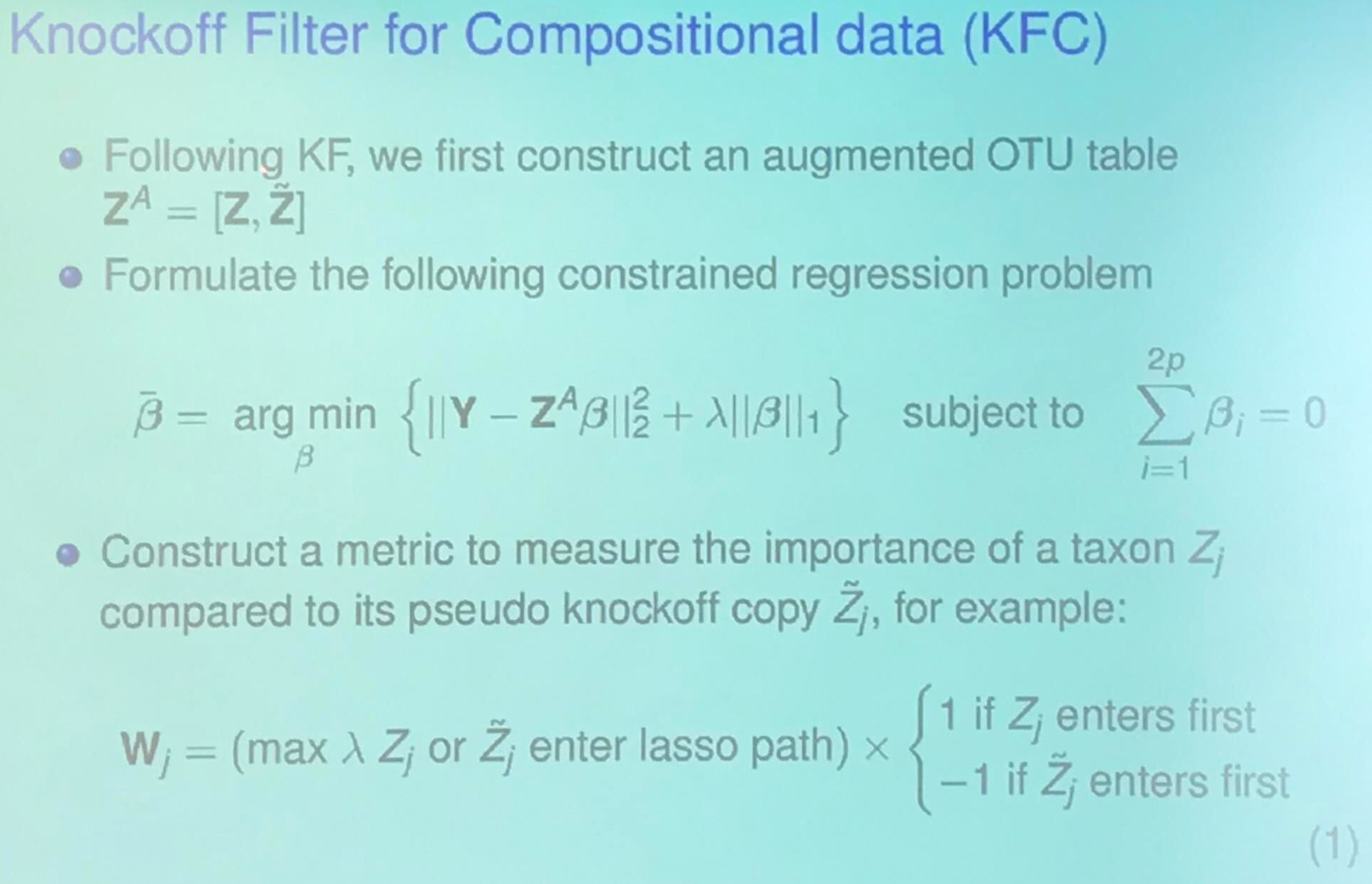
Knockoff Filter