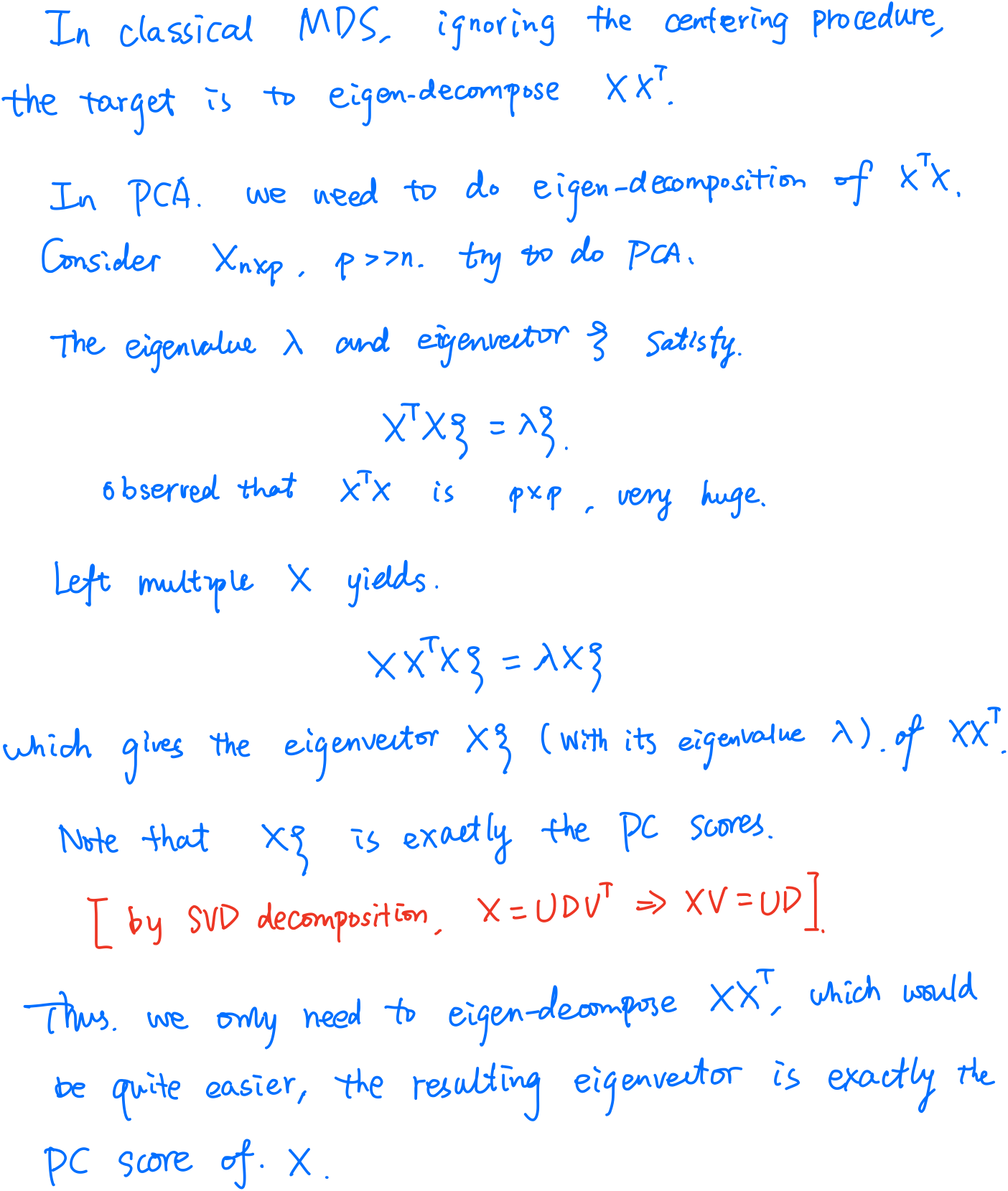Generalized Matrix Decomposition
Posted on (Update: )
This post is based on the talk given by Dr. Yue Wang at the Department of Statistics and Data Science, Southern University of Science and Technology on Jan. 04, 2020.
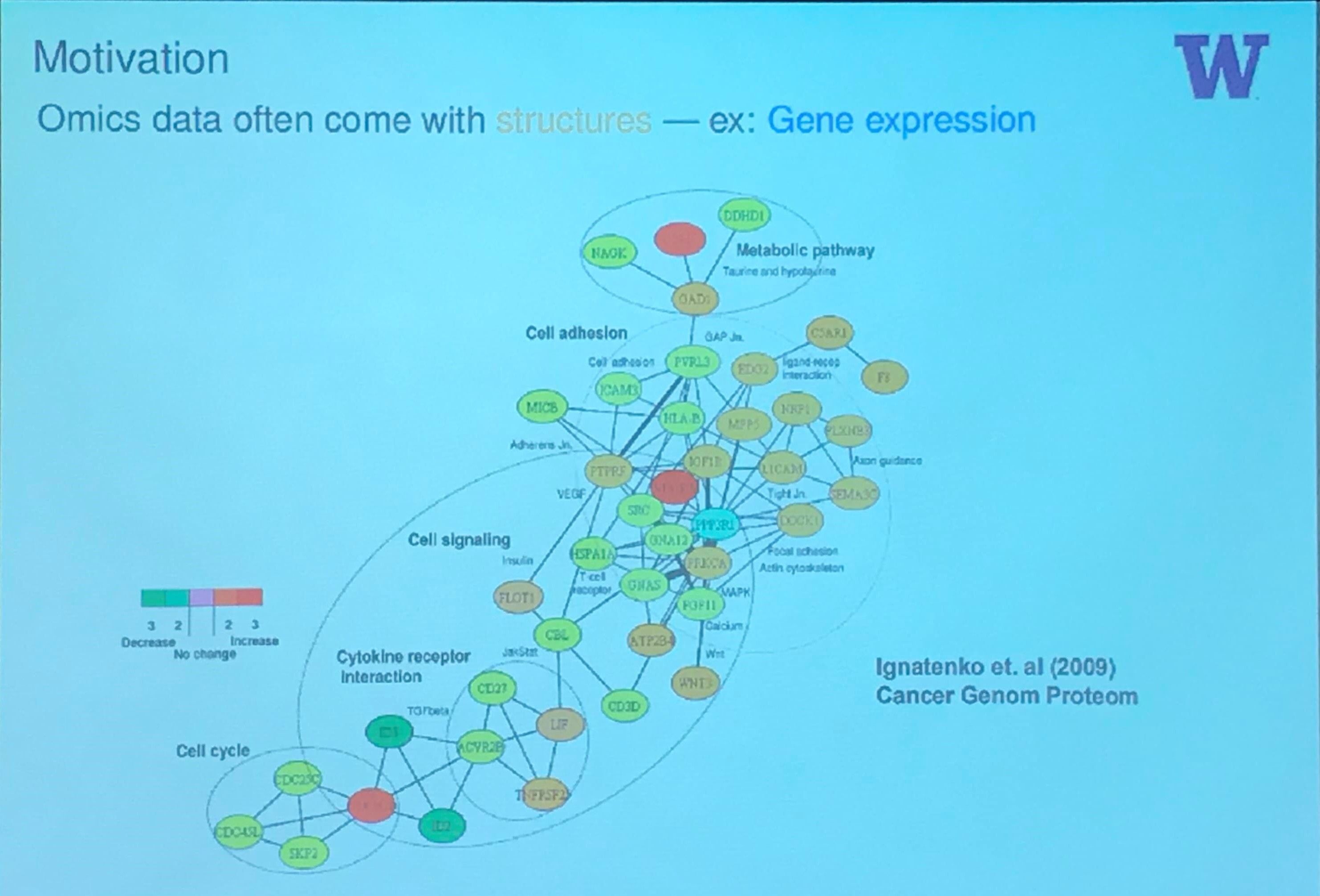
Motivation
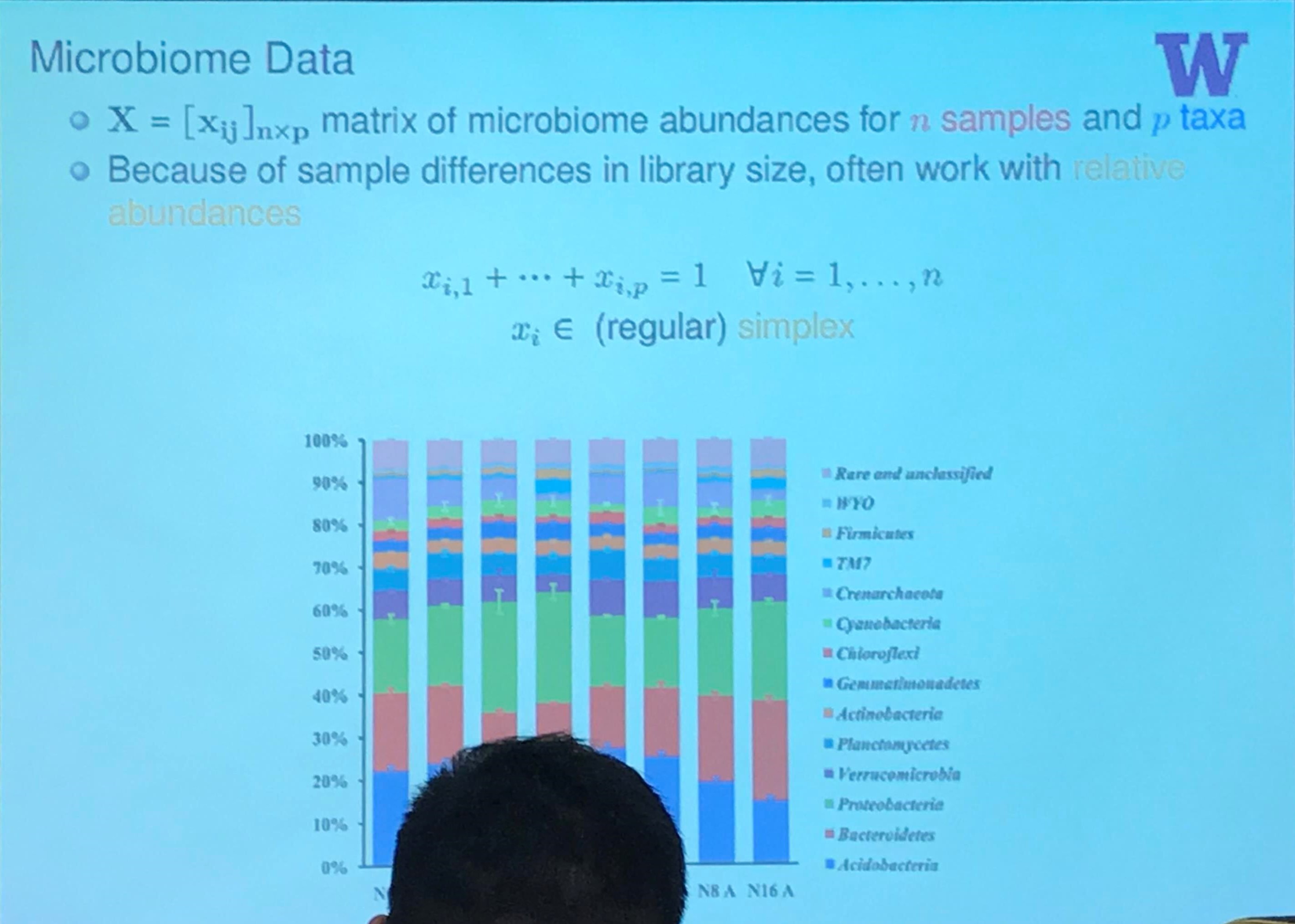
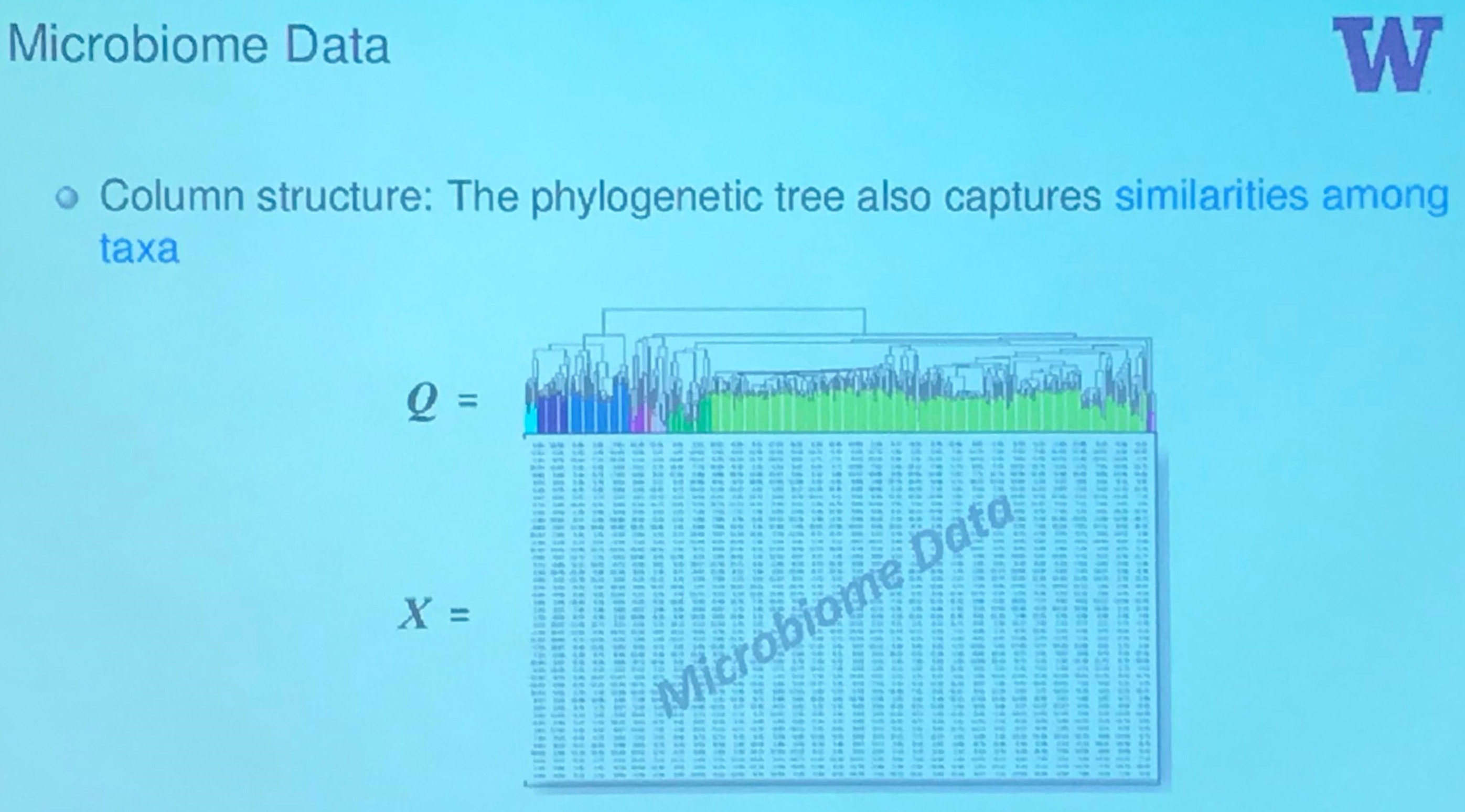
Microbiome Data
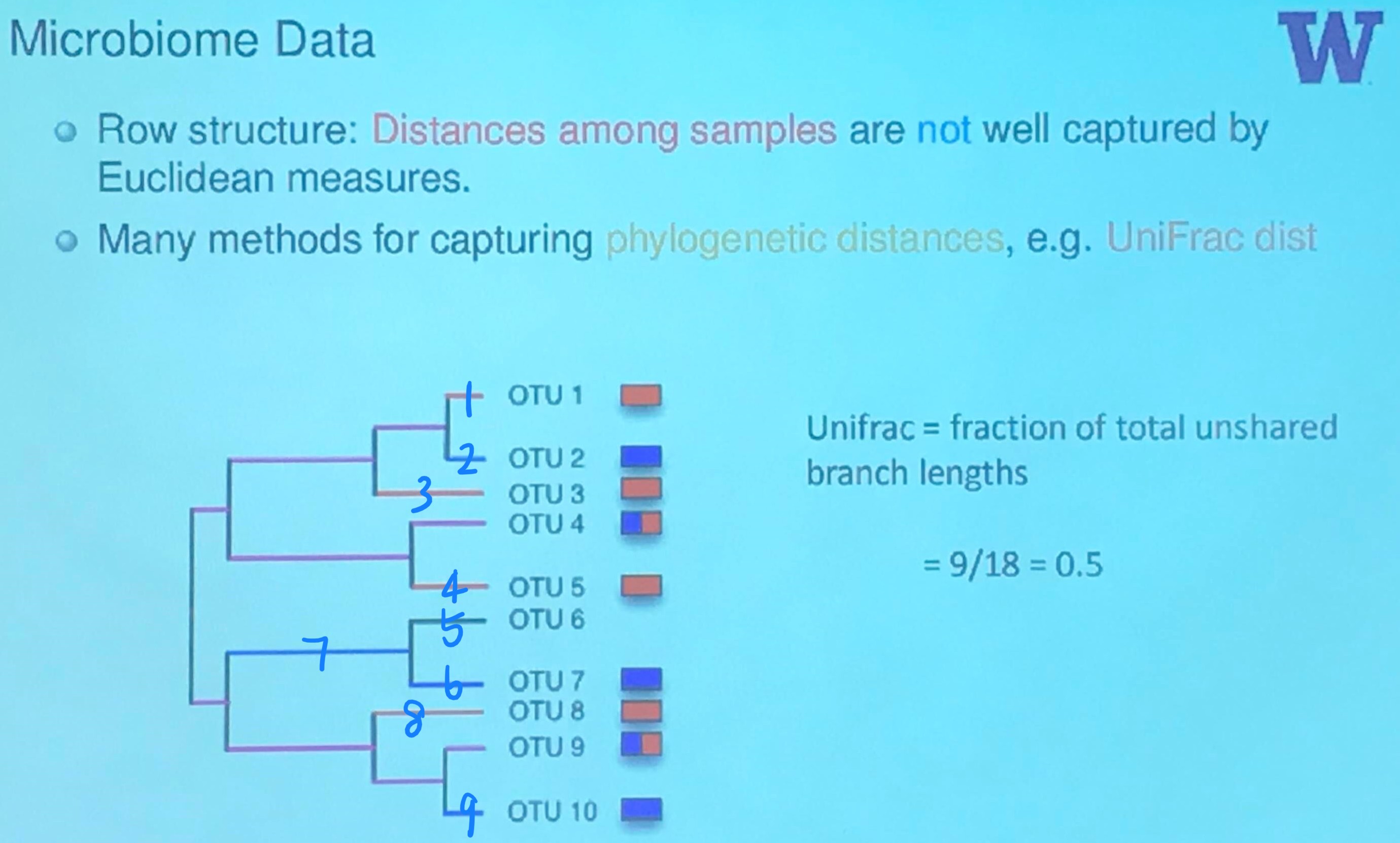
Here is a great tutorial with python code to calculate $\beta$ diversity, including the UniFrac, and Bray-Curtis.

Exploratory Analysis: Sample Clustering
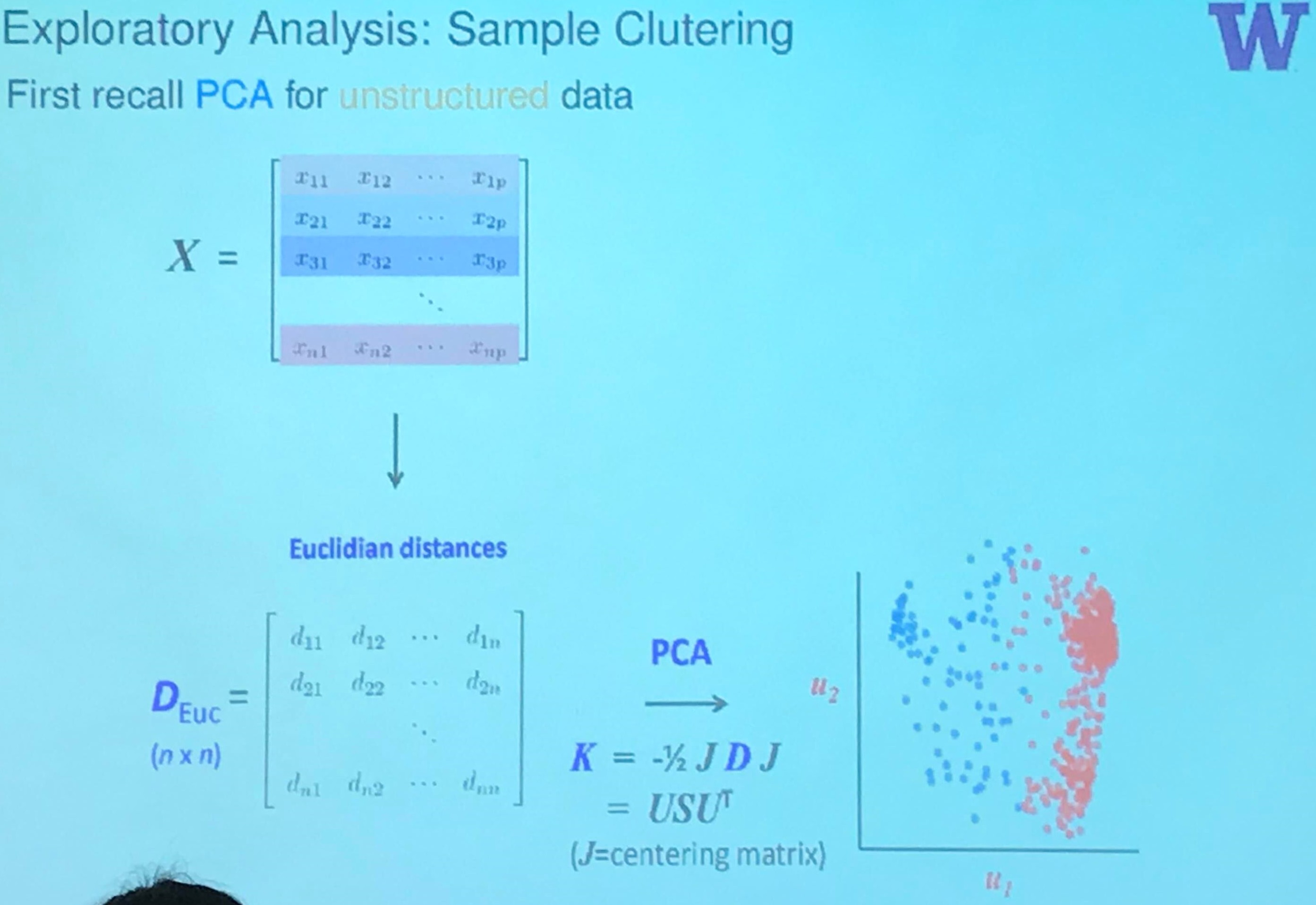
Actually, it should be treated as classical scaling, an approach of multidimensional scaling (MDS), but the classical scaling is equivalent to the principal analysis if the similarity is defined as the centered inner-products.
But here is still some differences. In the slide, it is the distance, the square root of the inner-product. And note that if $X=USV^T$, then $X^TX = VD^2V^T$ and $XX^T=UD^2U^T$.
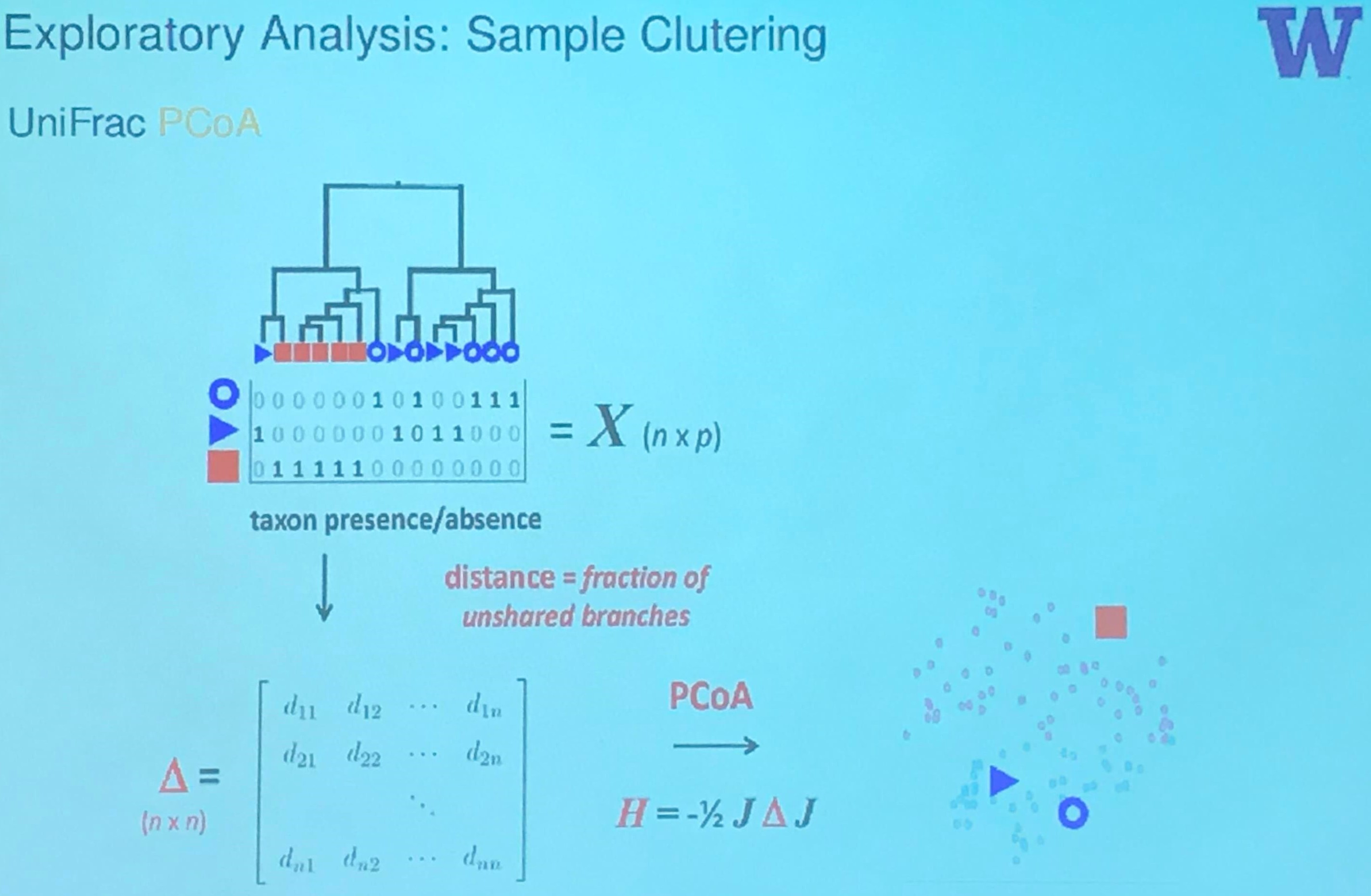
The wikipedia page of MDS says that MDS is also known as Principal Coordinate Analysis.
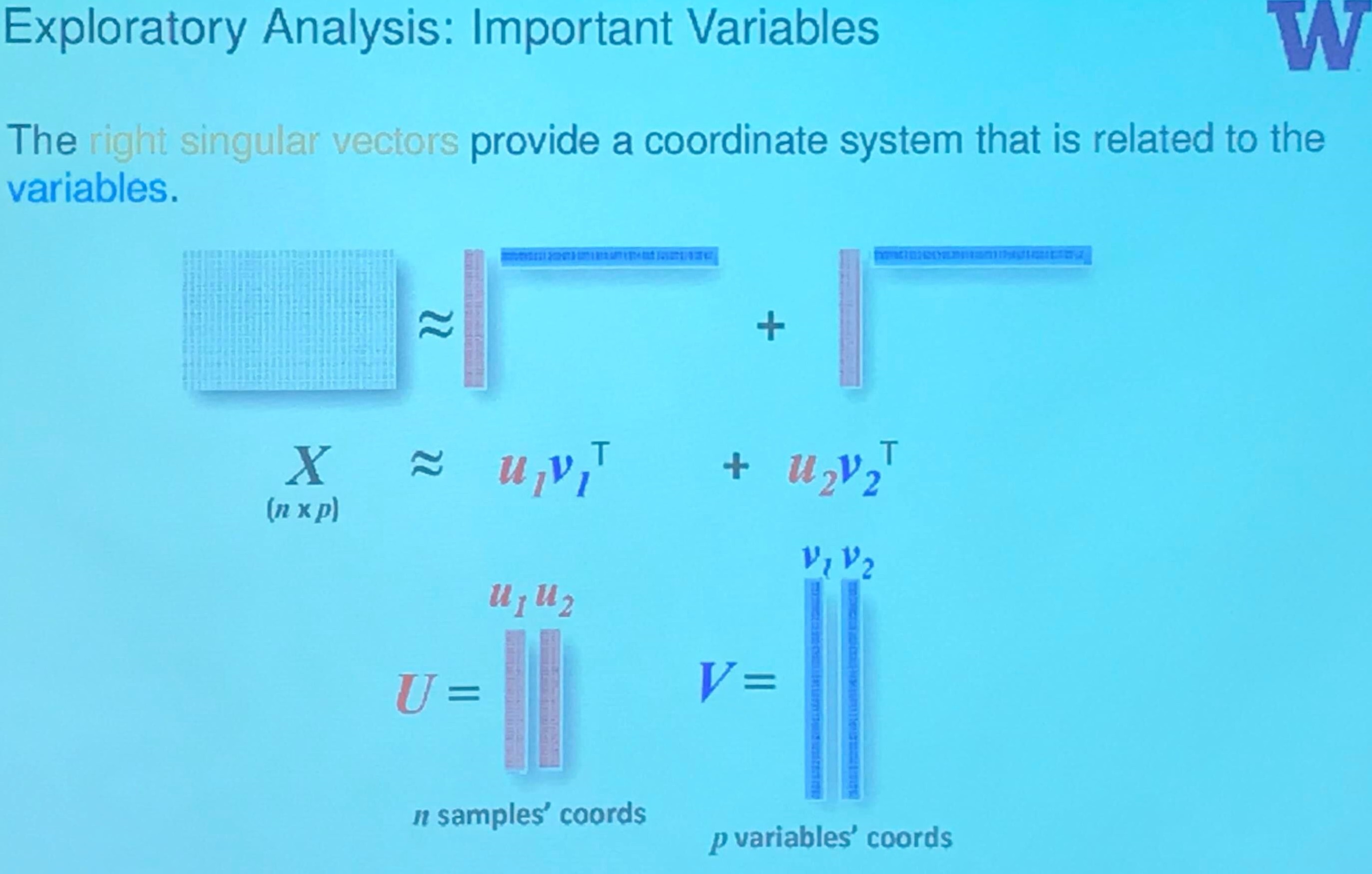
Exploratory Analysis: Important Variables
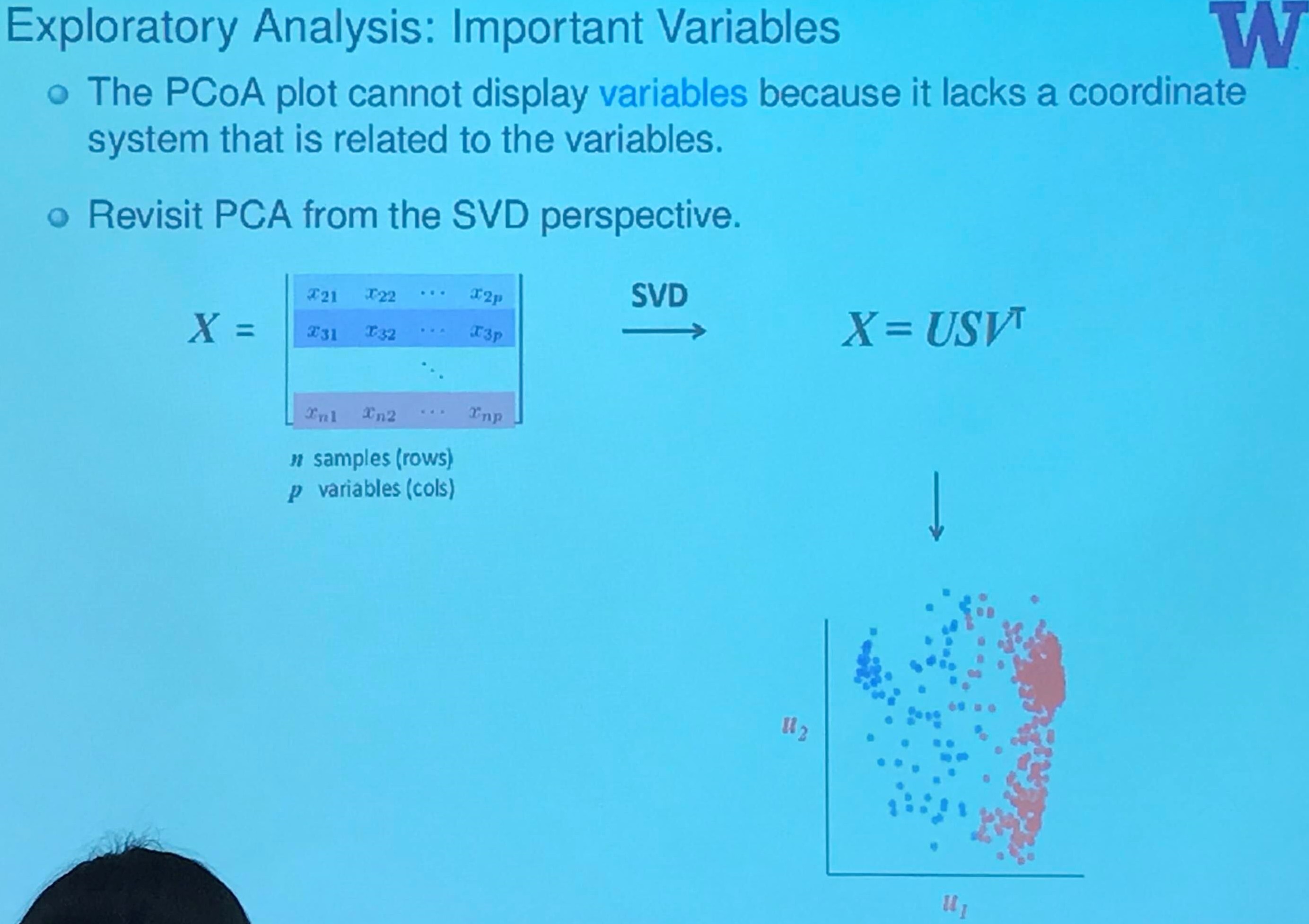
If we consider them in separate coordinate system respectively, then
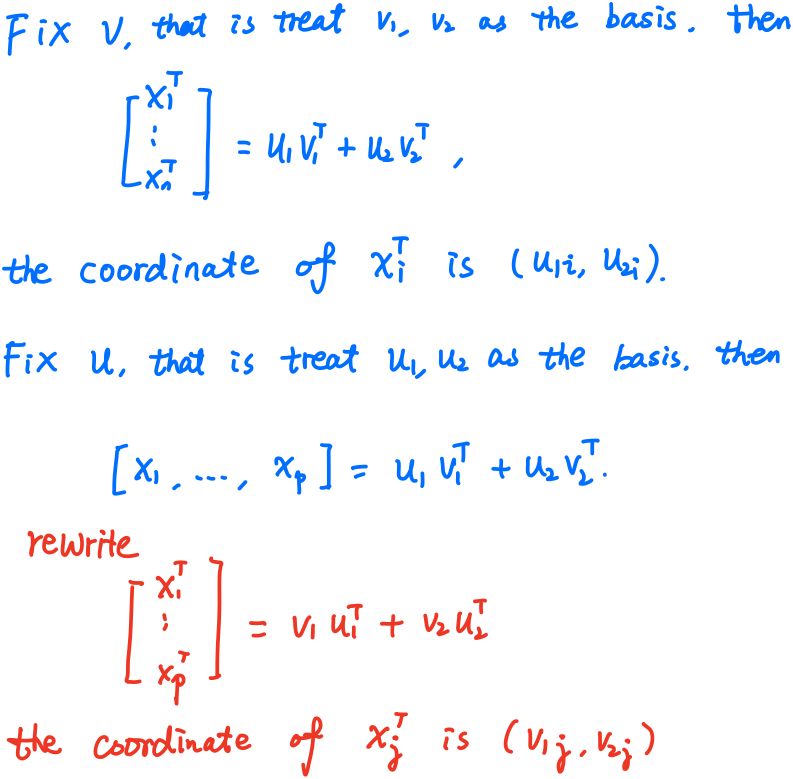
but if we put them into a single coordinate system, then
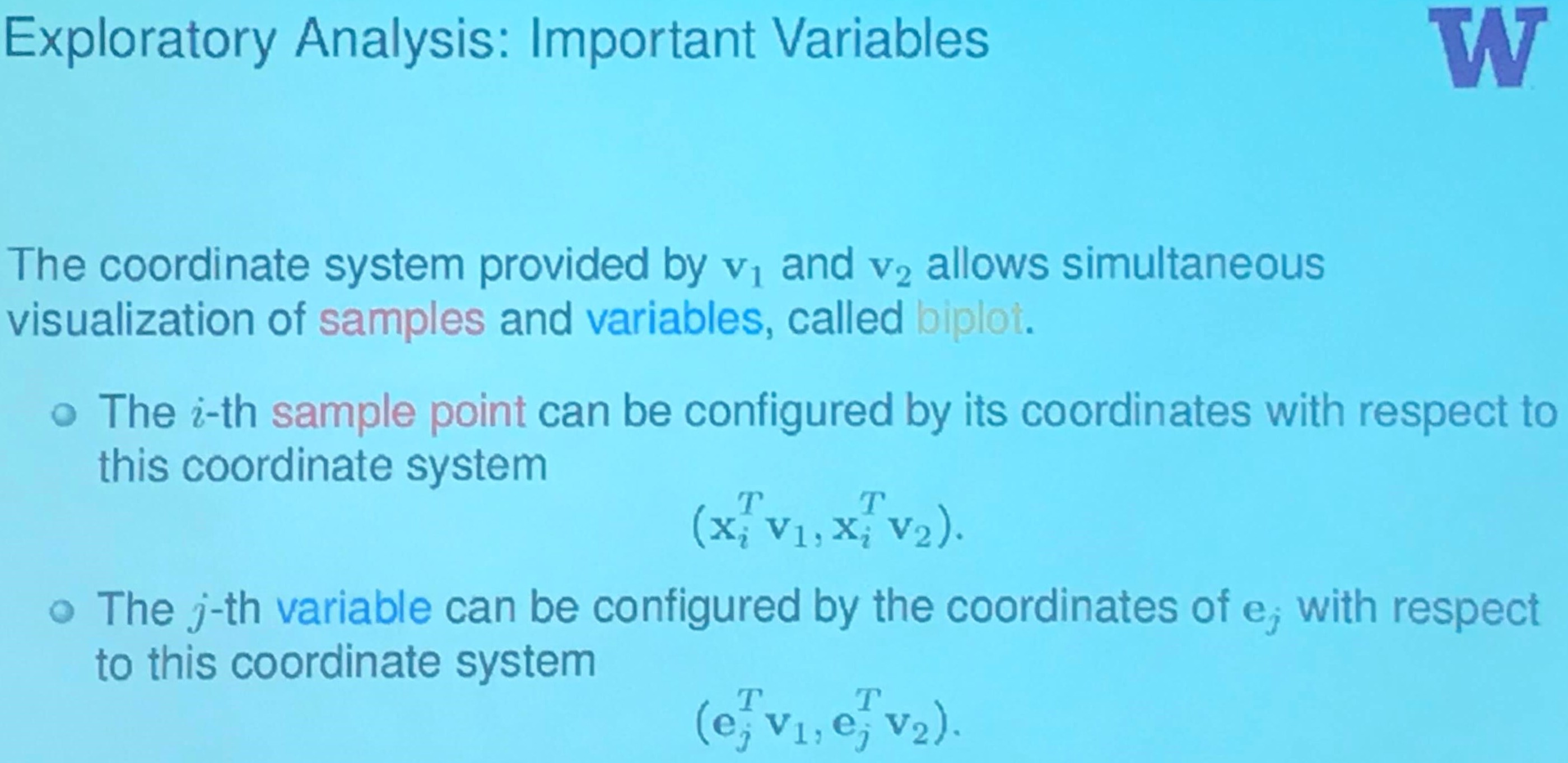
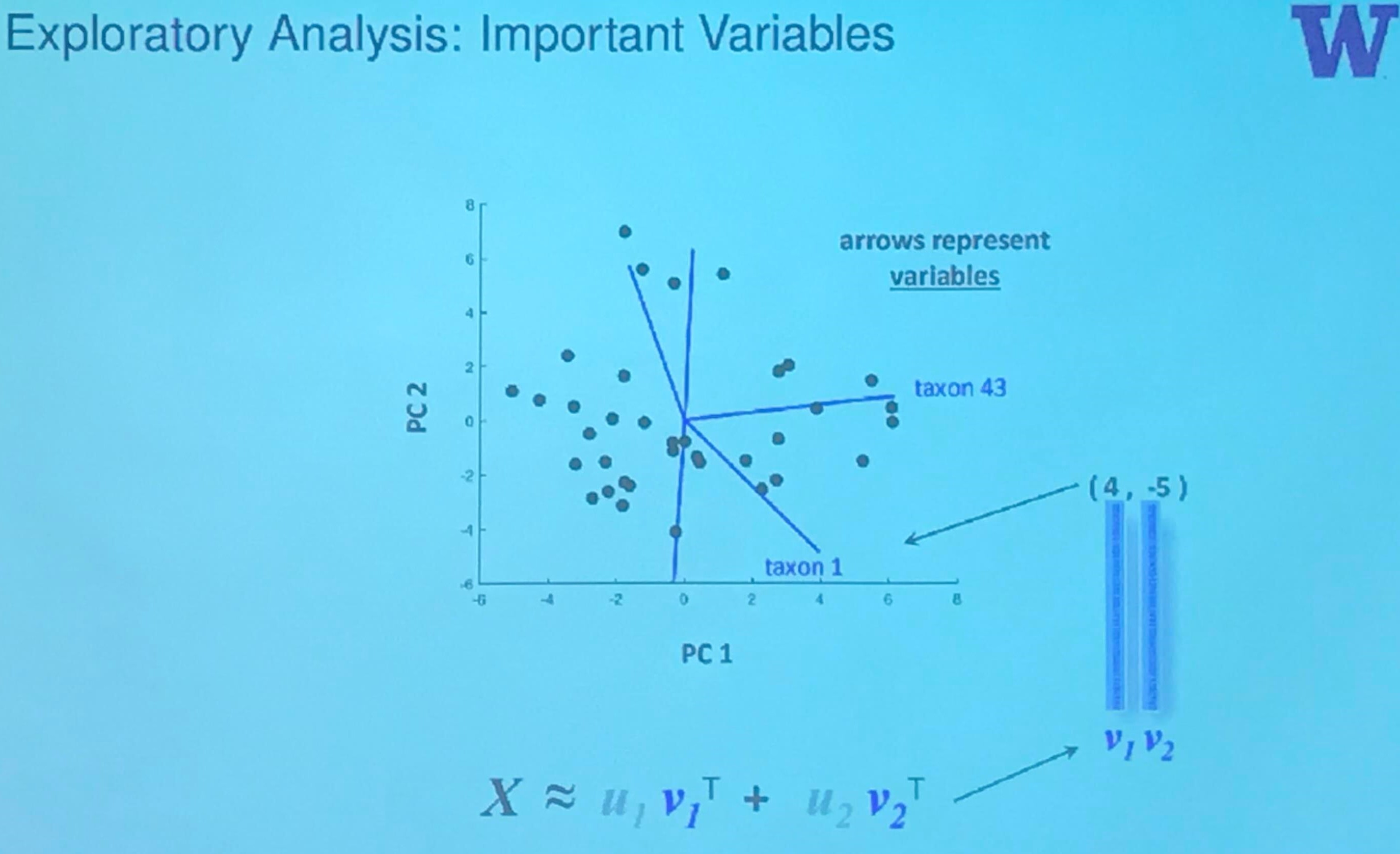
where the biplot is
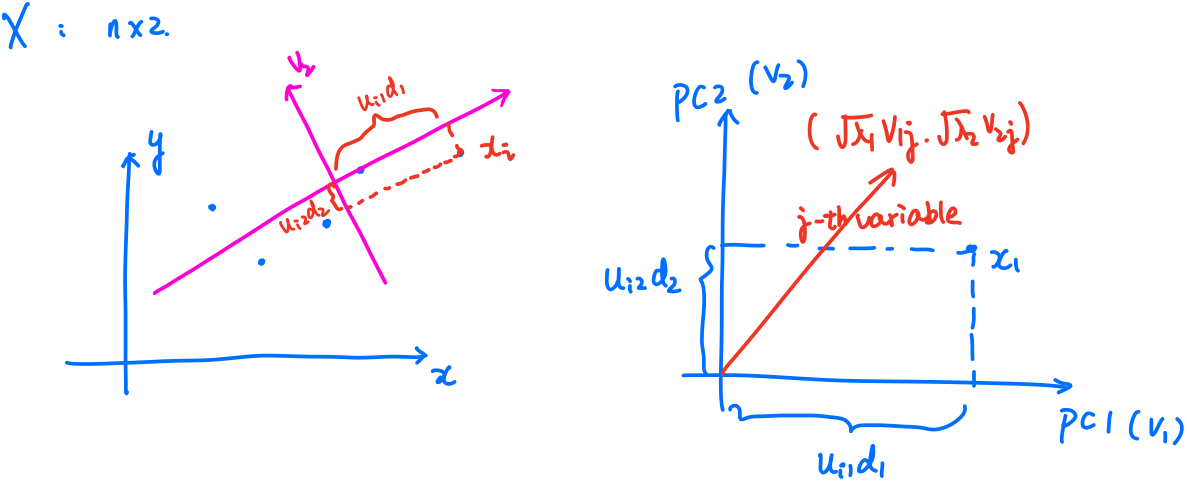
The GMD-biplot


Smokeless Tobacco Data

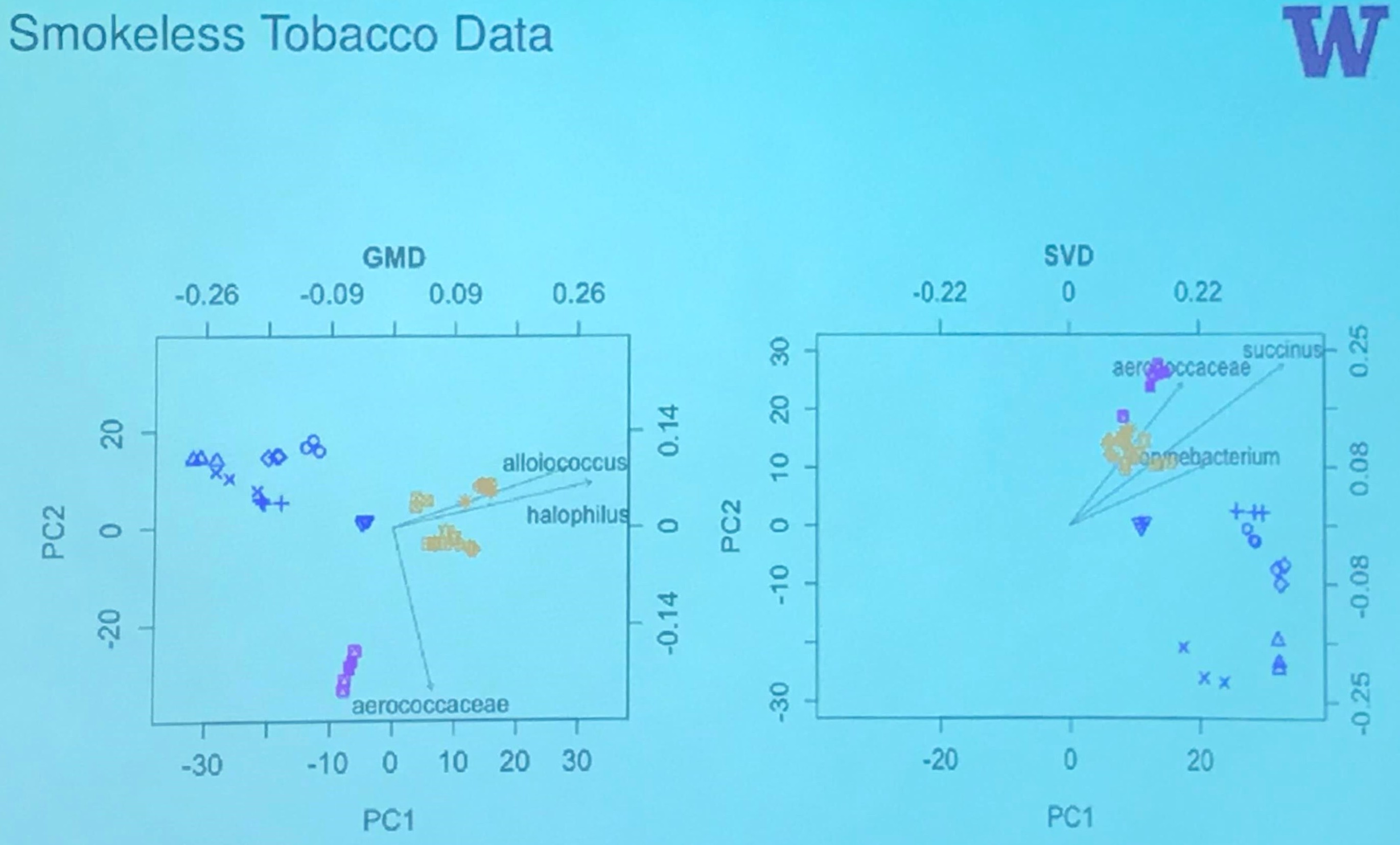
The author also writes a tutorial for the GMD-biplot.
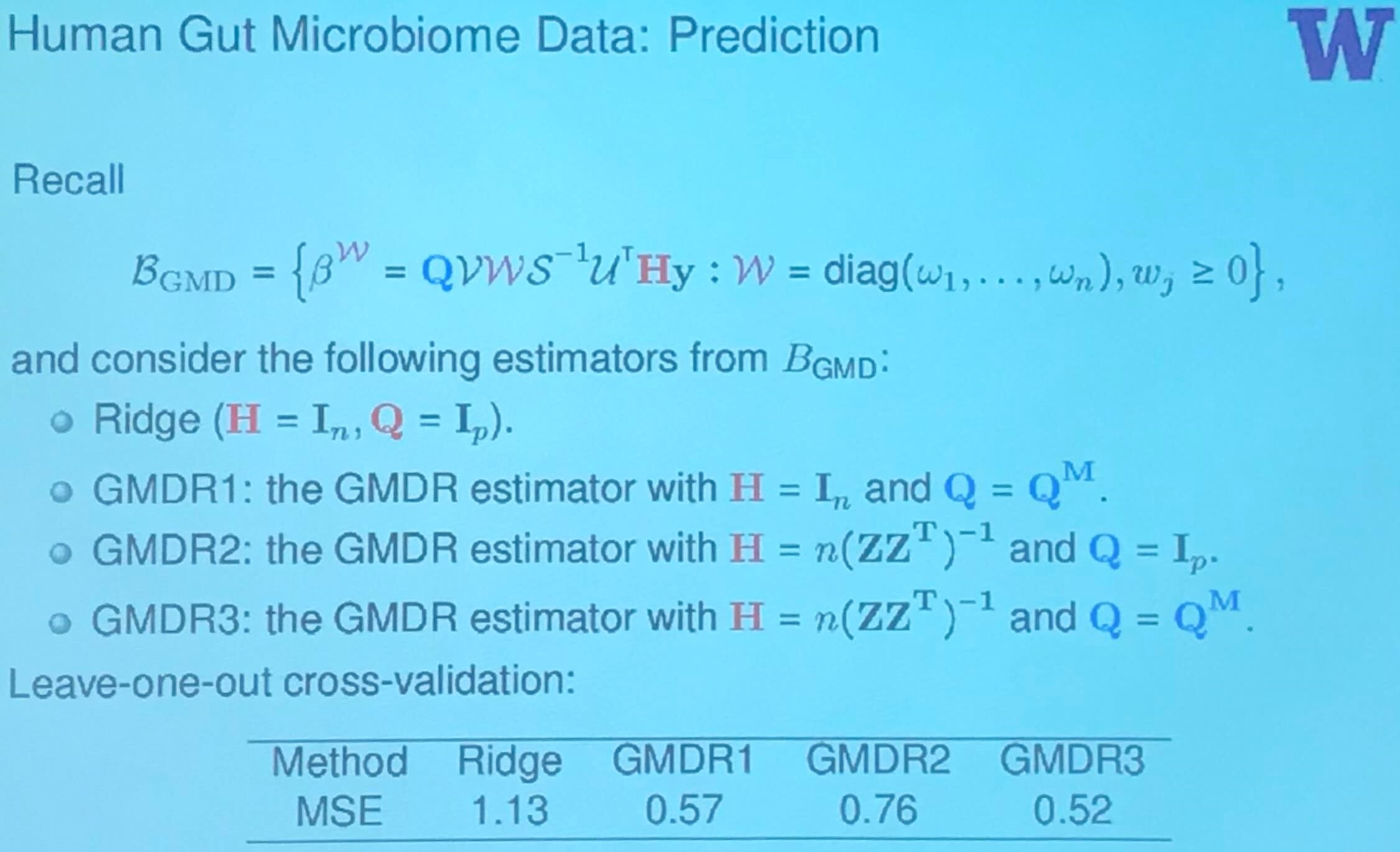
Supervised Learning with the GMD: GMDR


where the variable importance is calculated using the response.


why propose such a space of $\beta$? just want to exhibit the randomness of weights?
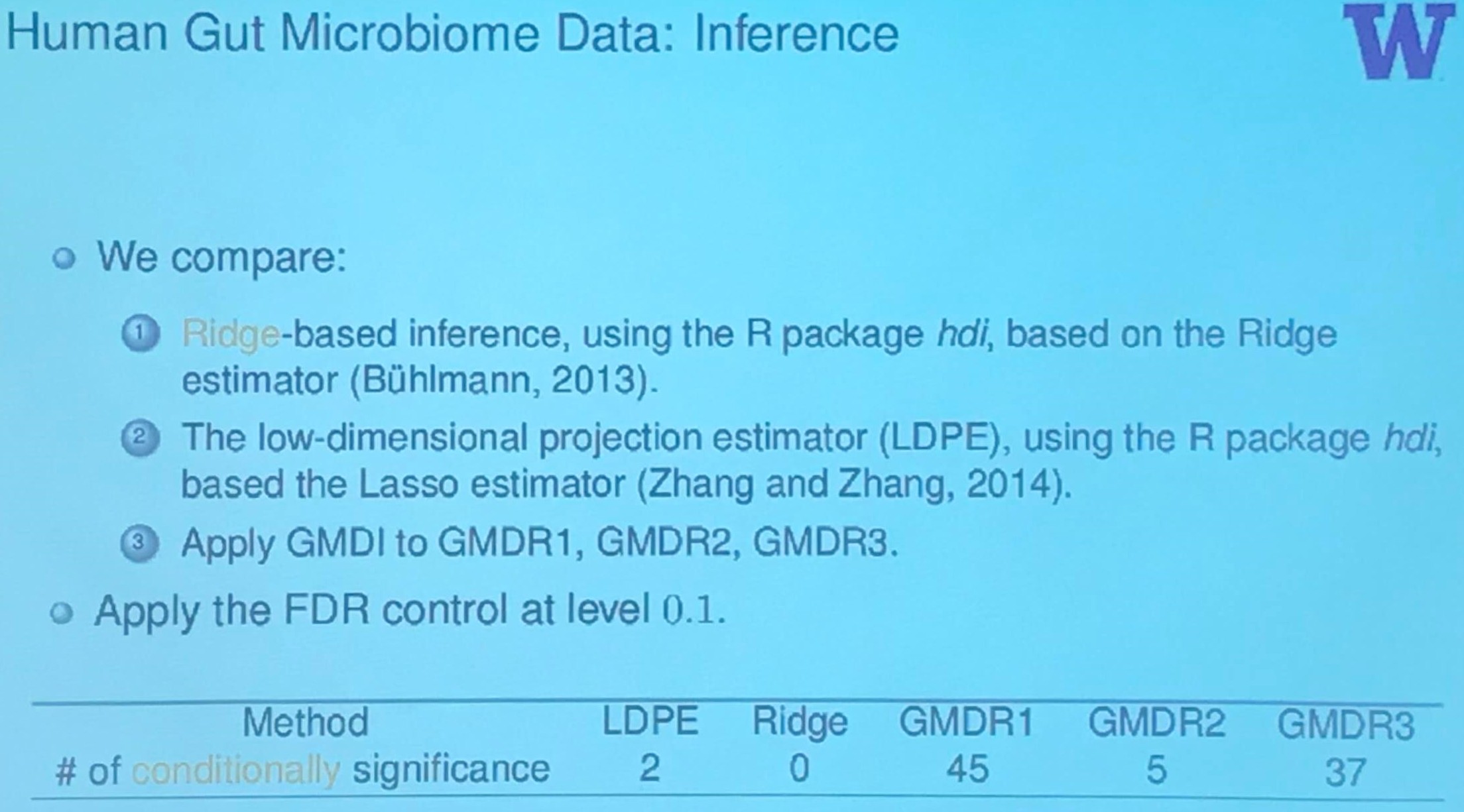
Inference




what is $D$ here?


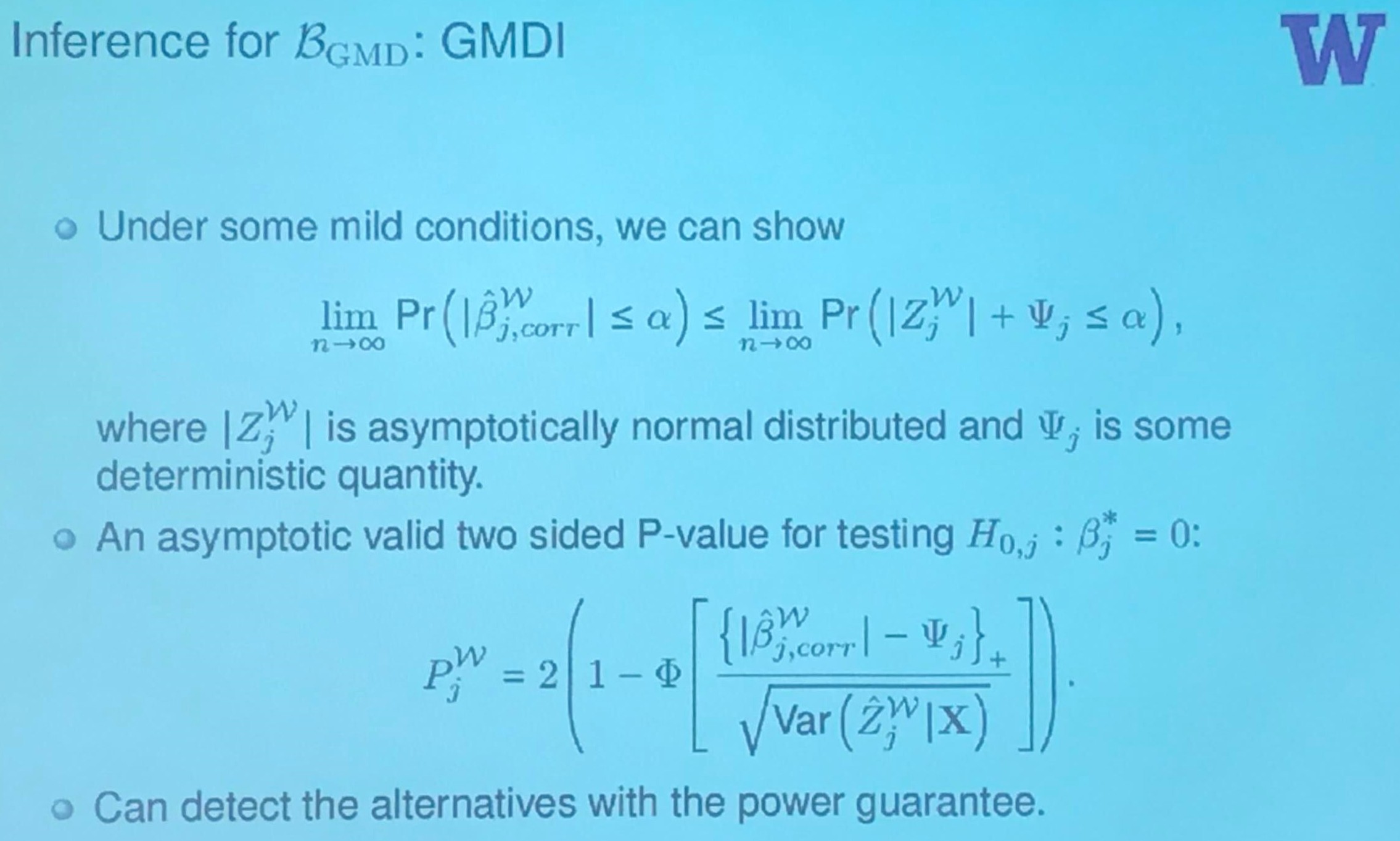
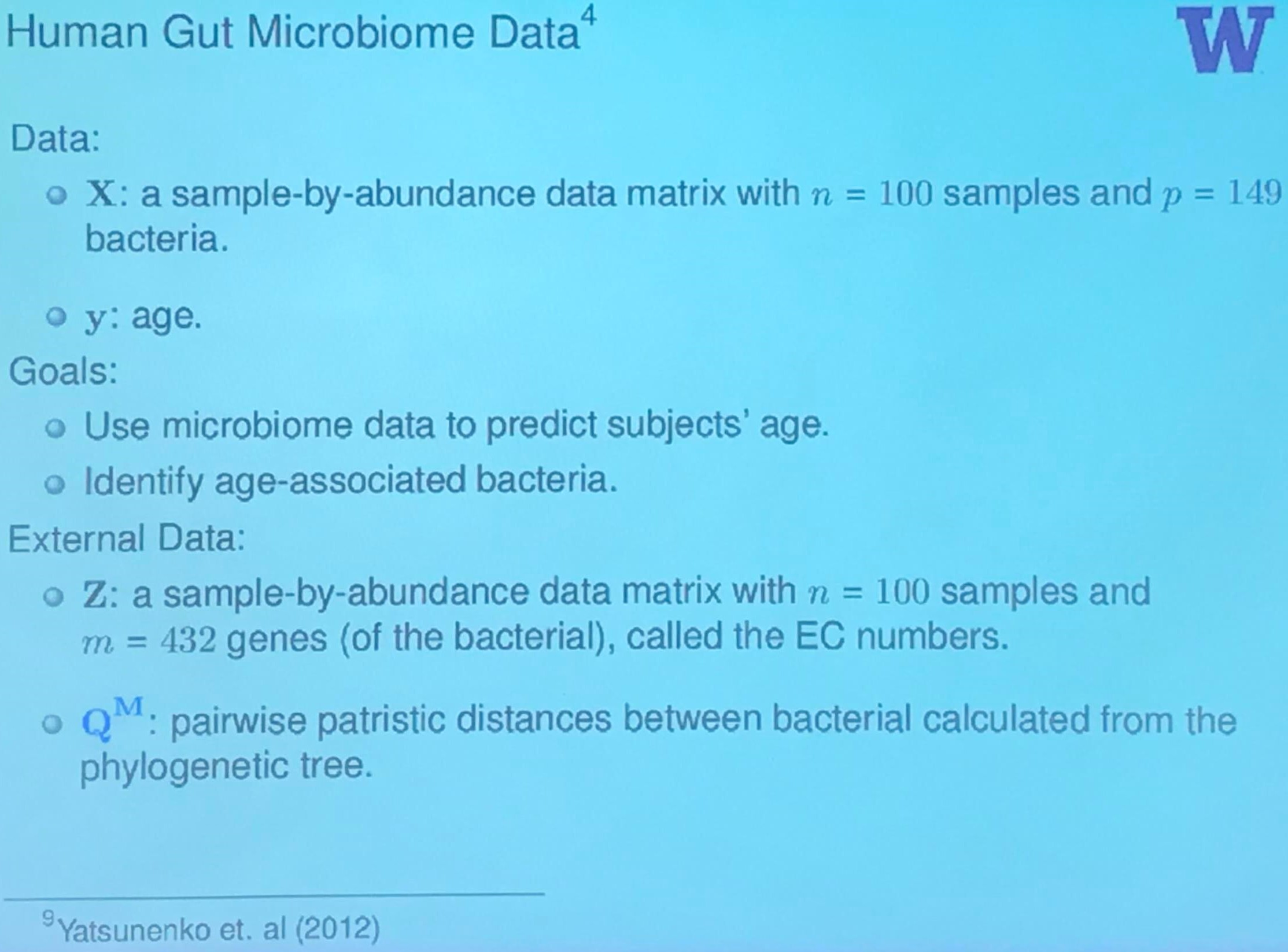
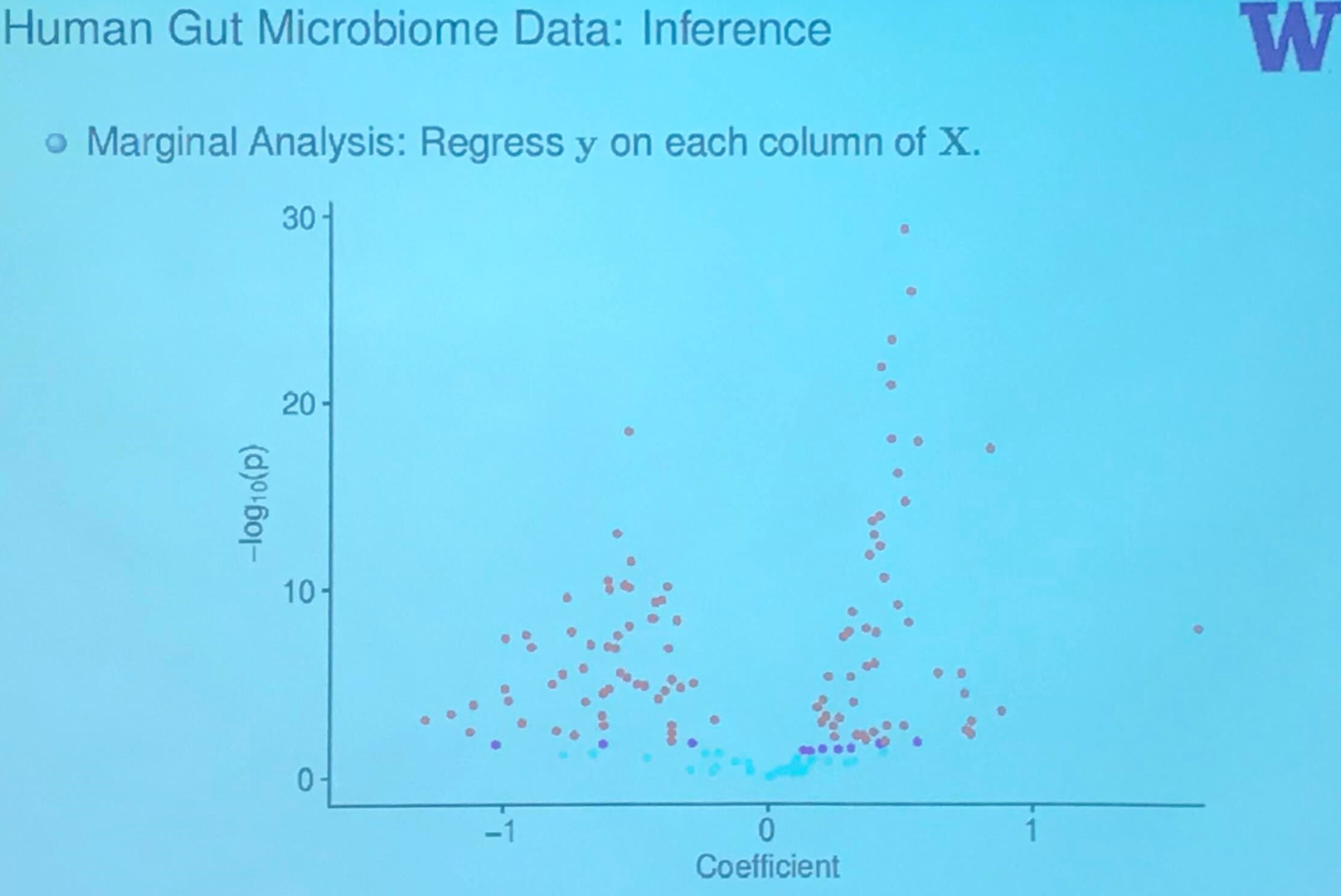
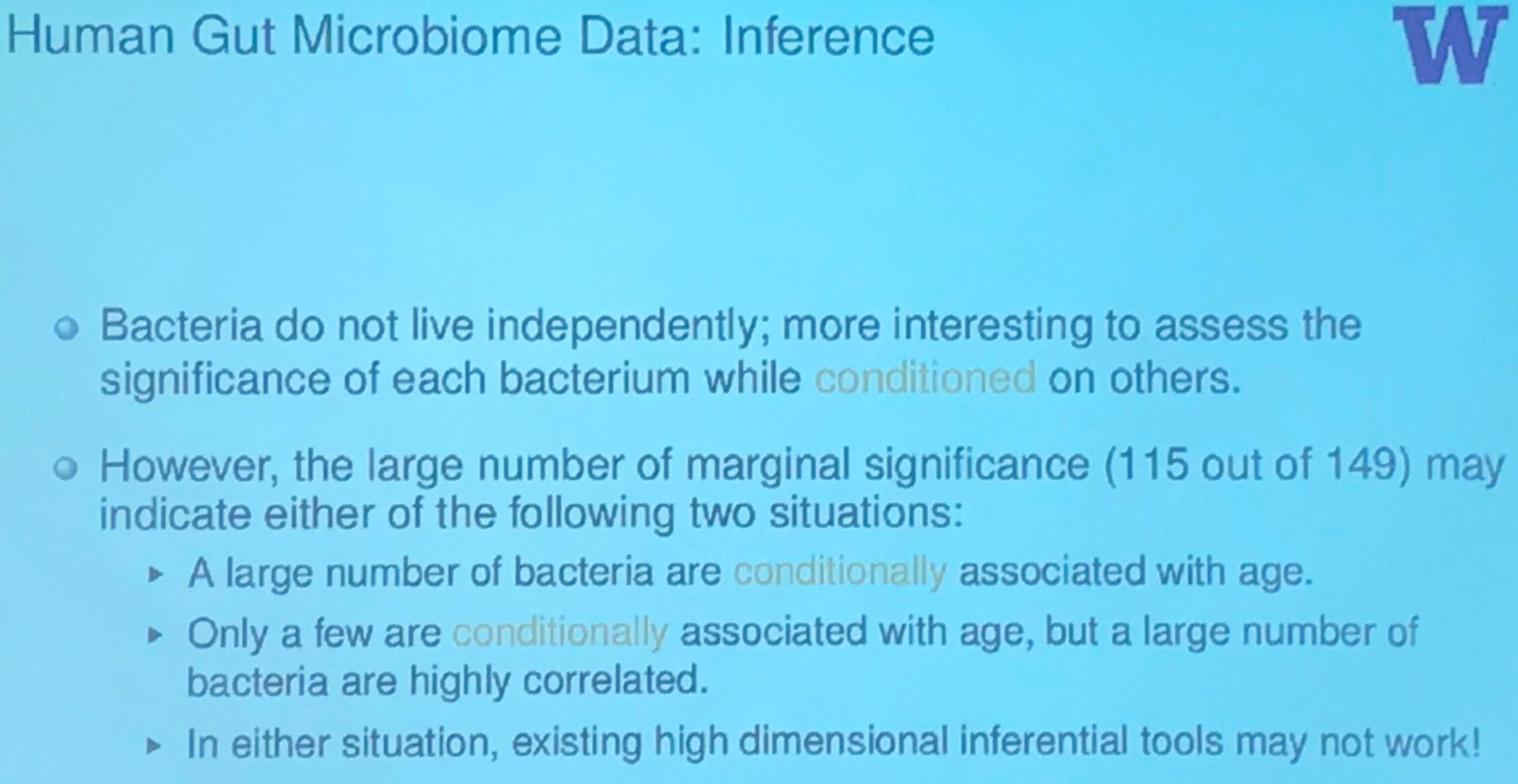
Human Gut Microbiome Data: Inference
Summary